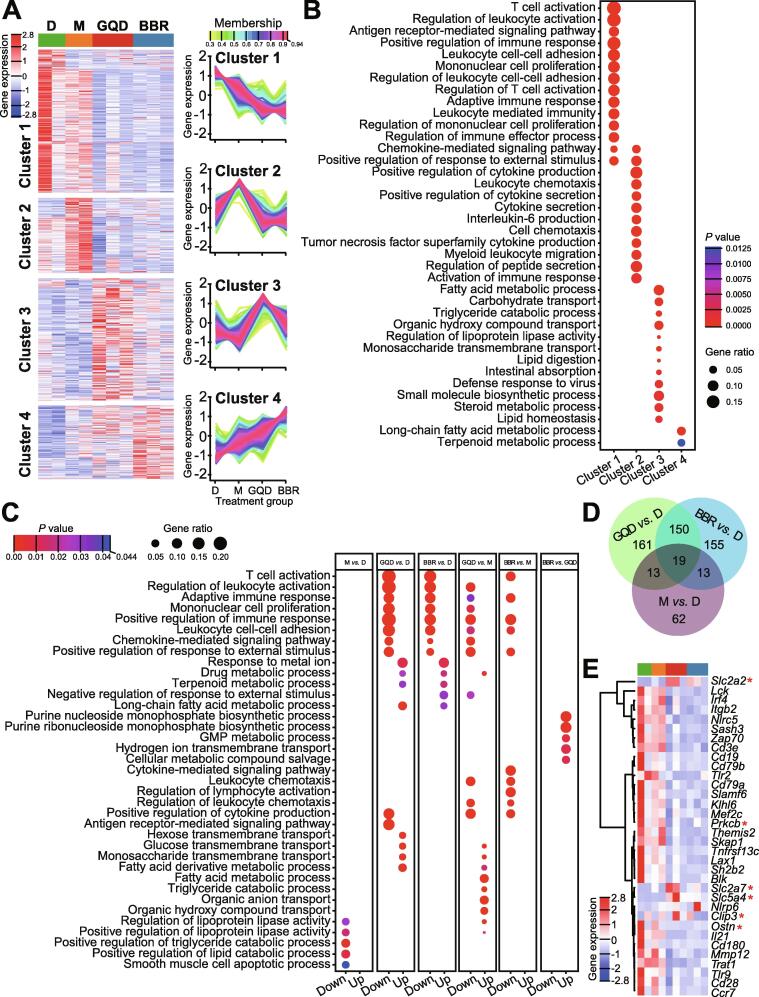
Figure 4.
Comparisons of gene expression profiles by RNA-seq among different treatment groups in the rat ileum
A. Genewise clustering heatmap of all 855 DEGs according to the gene expression patterns, showing segregation into four clusters. Cluster 1 (n = 295) includes genes with reduced expression in GQD and BBR treatment groups. Cluster 2 (n = 156) includes genes with expression upregulated in M group and downregulated in GQD and BBR groups. Cluster 3 (n = 252) includes genes with expression upregulated in both GQD and BBR groups, but higher in GQD group. Cluster 4 (n = 152) includes genes with increased expression among all three treatment groups, with the highest expression levels in BBR group. B. Doptplot of GO terms enriched for the four clusters. C. GO terms enriched among the upregulated DEGs and downregulated DEGs between the indicated groups (M vs. D; GQD vs. D; BBR vs. D; GQD vs. M; BBR vs. M; BBR vs. GQD). All P values in the GO enrichment analysis were adjusted for multiple testing using the BH method. Adjusted P < 0.05 was considered significant. Dot size represents the ratio of the number of DEGs to the number of genes in the corresponding entry. Dots are color-coded according to the adjusted P value for each term. D. Venn diagram displaying the shared DEGs between the three treatment groups. E. Heatmap of the representative DEGs that are shared by GQD vs. D and BBR vs. D comparisons and enriched for the GO terms “positive regulation of immune response” (GO:0050778) or “carbohydrate transport” (GO:0008643, denoted with a red asterisk), including immune and metabolic genes. DEG, differentially expressed gene; GO, Gene Ontology.