Abstract
The Phytophtora root and stem rot is a serious disease in soybean. It is caused by the oomycete pathogen Phytophthora sojae. Growing Phytophthora resistant cultivars is the major method of controlling this disease. Resistance is race- or gene-specific; a single gene confers immunity against only a subset of the P. sojae isolates. Unfortunately, rapid evolution of new Phytophthora sojae virulent pathotypes limits the effectiveness of an Rps (“resistance to Phytophthora sojae”) gene to 8–15 years. The current study was designed to investigate the effectiveness of Rps12 against a set of P. sojae isolates using recombinant inbred lines (RILs) that contain recombination break points in the Rps12 region. Our study revealed a unique Rps gene linked to the Rps12 locus. We named this novel gene as Rps13 that confers resistance against P. sojae isolate V13, which is virulent to recombinants that contains Rps12 but lack Rps13. The genetic distance between the two Rps genes is 4 cM. Our study revealed that two tightly linked functional Rps genes with distinct race-specificity provide broad-spectrum resistance in soybean. We report here the molecular markers for incorporating the broad-spectrum Phytophthora resistance conferred by the two Rps genes in commercial soybean cultivars.
Subject terms: Plant sciences, Plant breeding, Plant genetics
Introduction
Soybean [Glycine max (L.) Merr.] is one of the main oilseed crops produced and consumed worldwide and is among the world’s five utmost significant food crops1. Its production is limited by several soybean diseases, with an average annual yield loss of 11% in the United States2. Phytophthora root and stem and root rot (PRS) disease is in the top five diseases that cause severe annual yield losses in soybean3. The annual crop damage from PSR between 2003 and 2005 averaged about $251.6 million3,4. From 2010 to 2014, in 28 US states and Ontario, Canada, PSR caused an estimated annual yield loss of $403 millions3. The PSR disease in soybean is caused by the soil-borne oomycete Phytophthora sojae5,6, and soybean plants infected with P. sojae are more susceptible to infection by other soil-borne pathogens.
Oomycete pathogens are challenging to control and most fungicides are ineffective because the P. sojae infected roots are difficult to treat effectively with chemicals. Another difficulty is that many oomycetes appear to have extraordinary genetic flexibility, enabling them to adapt to and overcome rapidly the chemical control measures as well as host resistance mechanisms7,8. Methods employed to control PRS include fungicide applications9, planting resistant cultivars10,11, improvement in soil drainage8, modification of tillage practices12, and application of calcium-containing compounds13,14. The most effective way to reduce PRS damage is planting Phytophthora resistant soybean cultivars11.
Single dominant Rps genes confer resistance against P. sojae isolates that carry the cognate avirulence (Avr) gene. Soybean Rps genes activate effector-triggered immune responses15, as in other pathosystems16. More than 30 Rps genes/alleles have been mapped to nine chromosomes, including the newly identified Rps genes, RpsGZ and RpsX17,18. The Rps1 locus contains five functional alleles (Rps1a, 1b, 1c, 1d, and 1k)19–21, and the Rps3 locus contains three (Rps3a, 3b, and 3c)21,22. The Rps genes mapped to Chromosome 3 include Rps1, Rps7, Rps9, RpsYu25, RpsYD29, RpsYD25, RpsUN1, Rps1? and RpsWY14,20,23–28. While Rps2 gene and RpsUN2 have been mapped to Chromosome 1624,29,30, and the three Rps3 alleles, Rps3a, Rps3b, and Rps3c along with Rps8 and RpsSN10 to Chromosome 1322,24,31–33. The Rps4, Rps5, Rps6, Rps12, and RpsJS genes are tightly linked and are located on the lower arm of Chromosome 1817,24,34–36. Rps10 has been mapped to Chromosome 1723, RpsYB30, and RpsZS18 to Chromosome 1933,37 and Chromosome 237, respectively, and Rps11 to Chromosome 738.
P. sojae isolates evolve rapidly to overcome the introduced resistance genes in commercial cultivars, especially under the monoculture scenario. Over 200 known pathotypes of this pathogen have been reported and the number is ever growing presumably due to selection pressure on the P. sojae population for new pathotypes that can overcome the newly introduced Rps genes. The rapid evolution of new P. sojae virulent pathotypes limits the effectiveness of an Rps gene to 8–15 years39. For example, a survey on pathotype changes in the population of P. sojae over several decades showed that while 6% of the pathotypes could defeat the Rps1c gene from 1991 to 1994, it was 57% by 2004. While in 1994, no pathotype could defeat Rps1k, the number of pathotypes increased to 12% in 2004, and to 41% in 2015. The number of pathotypes that defeat both Rps1c and Rps1k increased from none to 31% between 1994 and 201539. With increased complexity of P. sojae pathotypes, new strategies for managing this pathogen are needed39. The use of resistant cultivars is the most cost-effective and environmentally safe method to control this disease. Henceforth, there is a constant need for novel Rps (“resistance to Phytophthora sojae”) genes to manage the disease effectively.
It was suggested that plant introduction (PI) line, PI399036 contains multiple Rps genes40,41. An Rps gene, Rps12, from this PI line was mapped to a 5.4 cM region between the simple sequence repeat (SSR) marker BARCSOYSSR_18_1840 and the NBSRps4/6-130/533 sequence17. To determine the utility of Rps12, we investigated the responses of recombinant inbred lines (RILs) containing Rps12 against a collection of P. sojae isolates. The objective of this study was to investigate the effectiveness of Rps12 against different P. sojae isolates. We utilized a set of recombinant inbred lines (RILs) containing recombination break points in the Rps12 region and lacking functional Rps genes in other known Rps loci. Investigation of the selected set of RILs for responses to a set of P. sojae isolates collected in Iowa revealed Rps13, linked tightly to Rps12 on Chromosome 18. The Rps12 and Rps13 genes together provide broad-spectrum Phytophthora resistance in soybean.
Materials and methods
Plant genetic material
The AX20925 RIL population used in this study was developed by crossing PI399036 with the germplasm line AR2. This population was used earlier to map Rps1217. The individual F2 plants were advanced to the F8 generation by applying the single-seed descent method41. In this study, 120 F8 families (recombinant inbred lines, RILs) were phenotyped for responses to a P. sojae isolate V13 that overcomes Rps12 encoded resistance and a mixture of isolates (R17 & Val12-11) that defeat Rps1a, 1b, 1c, 1d, 1k, 2, 3a, 3b, 3c, 4, 5, 6, and 7 genes. Of these 120 RILs, 60 were homozygous resistant, and 60 were homozygous susceptible. The 120 RILs were used in molecular mapping of the Rps13 gene.
Phytophthora sojae isolates
Phytophthora sojae isolates R17 (vir 1b, 1d, 3a, 3b, 3c, 5, 6), Val 12-11 (vir 1a, 1b, 1c, 1d, 1k, 2, 4, 7), 1005-2.9 (vir 1a, 1b, 1c, 1k, 3b, 7), III 5.2b (vir 1a, 1b, 1c, 1d, 1k, 7), III 23.4b (vir 1a, 1c, 1d, 2, 3b, 3c, 4, 5, 7), IV 5.2 (vir 1a, 1c, 1d, 2, 7), IV 10 (vir 1a, 1c, 1d, 7), IV 12.2a (vir 2, 4, 7), IV 13.4a (vir 7), IV 23.3 (vir 1a, 1c, 1d, 2, 7), VI 5.2b (vir 1a, 1c, 1d, 7), VI 12.1a (vir 6,7), VI 12.2b (vir 1d, 3a, 4, 5, 6, 7), VI 15 (vir 1d, 7), VI 20 (vir 1c, 2, 7), VI 23.3b (vir 1d, 7), PR1 (vir 7), PR6 (vir 7), S 5-5 (vir 7), V 13 (vir 1a, 1c, 1d, 4,7) and IV 6b (vir 1a, 1c, 1d, 7) were used for investigating the efficacy of Rps12 (Table 1). The P. sojae isolates were obtained from Anne Dorrance (Ohio State University, OH), Martin Chilvers (Michigan State University, MI), and Alison E. Robertson (Iowa State University). All isolates were grown on half-strength V8 agar plates amended with neomycin sulfate and chloramphenicol antibiotics for 5–7 days under room temperature in the dark as described by Dorrance et al. (2008)42.
Table 1.
Response of differential soybean lines and PI 399073 carrying Rps12 and Rps13 genes to 21 Phytophthora sojae isolates.
| Differential line | Rps gene | P. sojae isolates | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| III 5.2b | III 23.4b | IV 5.2 | IV 6b | IV 10 | IV 12.2 a | IV 13.4a | IV 23.3 | V 13 | VI 5.2b | VI 12.1 a | VI 12.2b | VI 15 | VI 20 | VI 17 | VI 23.3b | S 5-5 | R17 | Val 12-11 | 1005-2.9 | PR1 | ||
| L88-8470 | 1a | S | S | R | S | S | R | R | S | S | S | R | R | R | R | R | R | R | R | S | S | R |
| L77-1863 | 1b | S | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | S | S | R |
| Williams 79 | 1c | S | S | S | S | S | R | R | S | S | S | R | R | R | S | R | R | R | R | S | S | R |
| L93-3312 | 1d | S | S | S | S | S | R | R | S | S | S | R | S | S | R | R | S | R | S | S | R | R |
| Williams 82 | 1k | S | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | S | R |
| L82-1449 | 2 | R | S | R | R | R | S | R | S | R | R | R | R | R | S | R | R | R | R | S | R | R |
| L83-570 | 3a | R | R | R | R | R | R | R | R | R | R | R | S | R | R | R | R | R | S | R | R | R |
| L91-8347 | 3b | R | S | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | R | S | R |
| L92-7857 | 3c | R | S | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | S | R | R | R |
| L85-2352 | 4 | R | S | R | R | R | S | R | R | S | R | R | S | R | R | R | R | R | R | S | R | R |
| L85-3059 | 5 | R | S | R | R | R | R | R | R | R | R | R | S | R | R | R | R | R | S | R | R | R |
| L89-1581 | 6 | R | R | R | R | R | R | R | R | R | R | S | S | R | R | R | R | R | S | R | R | R |
| L93-3258 | 7 | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | R | S | S | S |
| PI 399073 | 8 | R | R | R | R | R | R | R | R | S | R | R | R | R | S | R | R | R | R | R | R | R |
Plants were rated seven days after inoculation as either R (resistant, < 30% seedling death) or S (susceptible, ≥ 70% seedling death).
Evaluation of genetic materials for Phytophthora resistance
Hypocotyls of 7-day-old seedlings of 120 RILs, the parents PI399036 and AR2 along with 14 differential lines carrying Rps1a, Rps1b, Rps1c, Rps1d, Rps1k, Rps2, Rps3a, Rps3b, Rps3c, Rps4, Rps5, Rps6, Rps7, and Rps8 genes and the susceptible cultivar ‘Sloan’ with no known Rps genes17,42–44 were inoculated using the wounded-hypocotyl inoculation technique45,46. The experiment was conducted three times. Plants were rated seven days after inoculation as either R (resistant, < 30% seedling death) or S (susceptible, ≥ 70% seedling death). Inocula were prepared using a modified version of the protocol described by Dorrance et al. (2008)42. The macerated R17 and Val 12–11 cultures were mixed in equal proportion to prepare the mixed inoculum45 that is virulent to soybean cultivars carrying Rps genes mapped to Rps1 to 7 loci and partially virulent to lines carrying Rps8. P. sojae strain V13 was also used as a separate inoculum as it is virulent to soybean lines carrying any of Rps1a, 1c, 1d, 4, 7, and 12 genes.
DNA preparation, bulked segregant analysis (BSA)
Before inoculation, one unifoliate leaf from each of 11 random plants of individual RIL was collected, bulked and frozen in liquid nitrogen, and stored at − 80 °C. The genomic DNA was extracted from the bulked leaf samples using the CTAB (cetyl trimethyl-ammonium bromide) method47. The identified SSR markers linked to Rps12 locus17 were used to conduct BSA for the Rps13 region on pooled DNA samples of 10 homozygous resistant RILs (Resistant Bulk) or ten susceptible RILs (Susceptible Bulk)17,48. In BSA48, a polymorphic molecular marker linked tightly to a target locus shows its allelic segregation either in coupling or repulsion phase linkage with alleles of the target locus. In BSA assays, the markers that are not linked to the target locus show heterozygosity due to recombination of the marker alleles with alleles of the target locus.
PacBio long-read sequencing and development of sequence-based polymorphic (SBP) molecular markers
A ~ 50 genome equivalents genome sequence of the PI399036 and AR2 was obtained by PacBio long-read sequencing at the DNA Facility, Iowa State University. The bowtie program was run to identify single nucleotide polymorphisms (SNPs) between genomes of the resistant (PI399046) and susceptible (AR2) lines by mapping sequence reads onto the 8 Mbp region spanning the 53–61 Mbp physical locations on Chromosome 18 containing Rps12 and Rps13. Over 26,000 putative SNPs were identified. We used SNPs of the putative Rps13 region to develop necessary SBP markers according to Sahu et al.49 for mapping the Rps13 gene. Among the identified SNPs, we looked for the ones that are polymorphic for restriction endonucleases. Polymerase chain termination reaction (PCR) amplicons of approximately 200 nucleotides DNA containing variations for restriction endonuclease sites between PI399036 and AR2 were considered as putative SBP markers. Finally, primers for PCR amplification were designed in such a way that one can easily distinguish the haplotype-specific restriction fragment length polymorphisms by separating the restriction enzyme digested PCR products on a 4% (w/v) agarose gel49. Seventeen SBP markers were identified for the Rps12-Rps13 region (Table S1).
Simple sequence repeats (SSR) and SBP markers were used to construct a linkage map of the genomic region carrying the putative novel Rps13 gene. Molecular markers based on previously reported NBSRps4/6 sequence and SSR markers17, and newly developed SBP markers were used in mapping the Rps13 gene (Table S1). SSR markers linked to RpsJS were also used in mapping the Rps13 region34. Eleven polymorphic SSR markers, two previously reported NBSRps4/6 molecular markers along with the newly developed five SBP markers were used to map the Rps13 gene17 (Tables S1, S2).
Screening RILs and parental lines for the presence of known Rps genes
Twenty-three SSR markers linked to the Rps1, 2, 3, 7, 8, 9, 10, 11, Yu25, WY, Rps1?, RpsUN1, UN2, and YD29 loci were used to evaluate for possible polymorphisms between the AR2 (susceptible), and PI399036 (resistant) parents (Table 2) in order to identify RILs that carry SSR alleles specific to the P. sojae susceptible AR2 parent.
Table 2.
SSR markers linked to known Rps regions.
| Rps gene | Linked SSR markers | Chromosome | Molecular linkage group |
|---|---|---|---|
| Rps1a, b, c, d, k | Satt152, Sat_186, Satt631, Satt683, Satt159, Satt530 & Satt009 | 3 | N (Gordon et al.40; Sugimoto et al.14; Wu et al.28; Sun et al.27; Lin et al.29) |
| Rps2 | Sat_144 & Satt440 | 16 | J (Gordon et al.40) |
| Rps3a, b, c | Satt335 & Satt510 | 13 | F (Gordon et al.40) |
| Rps4 | Sat_064 | 18 | G (Sandhu et al.35; Sahoo et al.17) |
| Rps6 | Sat_064 | 18 | G (Sandhu et al.35; Sahoo et al.17) |
| Rps7 | Satt631, Satt683, Satt152, Satt530 & Satt009 | 3 | N (Gordon et al.40; Sugimoto et al.14; Sun et al.27) |
| Rps8 | Satt663 | 13 | F (Gordon et al.40) |
| Rps9 | Satt631 & Sat186 | 3 | N (Wu et al.28; Lin et al.29) |
| Rps10 | Sattwd15-24, Sattwd15-25 & Sattwd15-47 | 17 | D2 (Zhang et al.23) |
| Rps11 | SSR_07_0286, SSR_07_0300 & SSR_07_0295 | 7 | M (Ping et al.38) |
| Rps12 | BARCSOYSSR_18_1840 & Sat_064 | 18 | G (Sahoo et al.17) |
| UN1 | Satt159 & SSR_03_0250 | 3 | N (Lin et al.29) |
| RpsUN2 | SSR_16_1275 & Sat_144 | 16 | J (Lin et al.29) |
| RpsYu25 | Sat186 & Satt152 | 3 | N (Sun et al.27; Lin et al.29) |
| YD29 | SattWM82-50 and Satt1k4b | 3 | N (Zhang et al.23) |
| RpsWY | Satt631 & Satt152 | 3 | N (Cheng et al.75) |
| Rps1? | Satt631, Sat186 & Satt009 | 3 | N (Sugimoto et al.14) |
| RpsJS | SSRG60684K & BARCSOYSSR_18_1861 | 18 | G (Sun et al.14) |
Linkage map construction and statistical analysis
The Chi-square (χ2) analysis was performed to check the phenotypic data for goodness-of-fit to a Mendelian segregation 1:1 ratio using Graphpad (http://www.graphpad.com/quickcalcs). Mapmaker version 3.050 and the Kosambi mapping function51 were used to calculate genetic distances in cM units from the recombination fractions between any given two loci. A logarithm of the odds (LOD) threshold was set as 3.0 to determine the linkages between studied loci. Mapmaker package uses the Lander-Green algorithm to calculate the “best” map order of loci50. The marker order was determined using the log-likelihood method50. The linkage map of molecular markers and the Rps genes was drawn using MapChart 2.352.
The source of Rps12 and Rpas13 genes
The PI399036 containing the two Rps genes, Rps12 and Rps13, is available from the USDA Soybean Germplasm Collection. The contact person for the seeds is Esther K Peregrine (esther.peregrine@ars.usda.gov), Assistant Soybean Curator, USDA/ARS SoybeanSoybean Germplasm Collection, 1101W. Peabody Dr., Rm. 180, National Soybean Research Center, Urbana, IL 61801, USA.
The segregating materials studied in this study were generated by author Silvia Cianzio and will be available from the Bhattacharyya lab, G319 Agronomy Hall, Iowa State University, Ames, IA 50011, USA. All plant collection methods were complied with relevant institutional, national, and international guidelines and legislation.
Results
Identification of putative RILs carrying the Rps12 gene
It was proposed that the Phytophthora resistant PI399036 line contains multiple Rps genes40. Earlier we mapped Rps12 of this line using a mixture isolates that overcome most known Rps genes17. To investigate the utility of Rps12 against a set of P. sojae isolates collected from Iowa soybean field, we looked for RILs that carry only Rps12. We have investigated 60 Phytophthora resistant RILs generated from the cross between PI399036 × AR217 for SSR markers linked to the known Rps regions as described below.
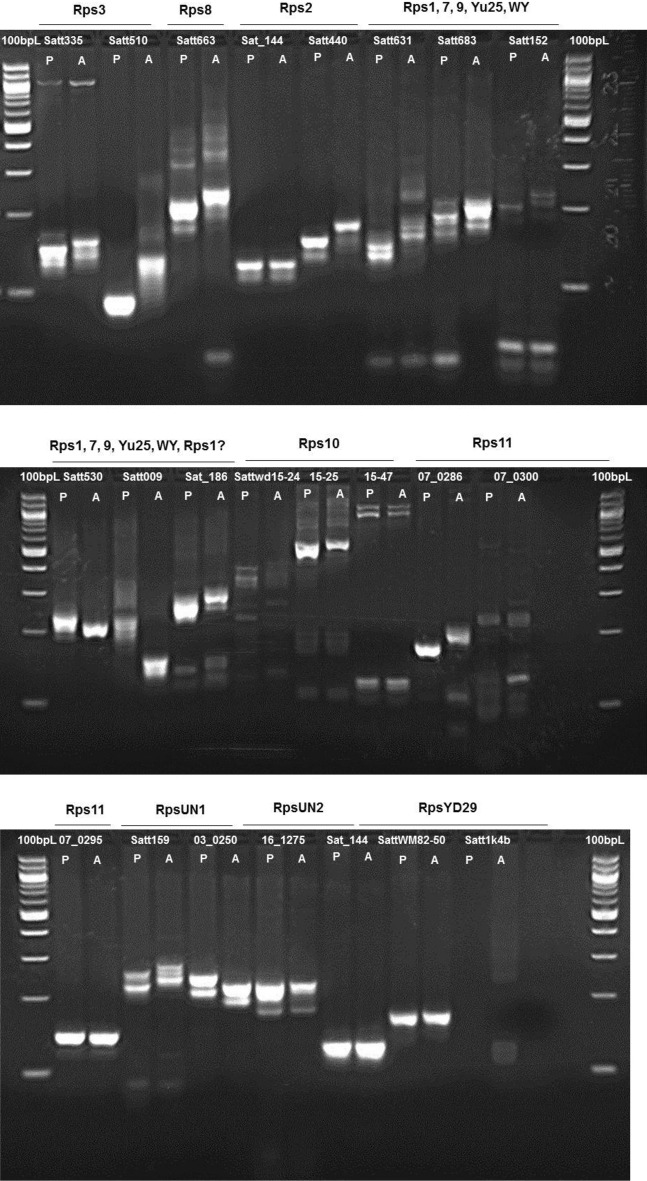
We used 23 SSR markers that were published earlier (Table 2). These include SSR markers Satt335 and Satt510 for Rps3 locus, Satt663 for Rps8 locus, Sat_144 and Satt440 for Rps2 locus, and Satt631, Satt683, Satt152, Satt530, Satt009 and Sat186 for Rps1, 7, 9, Yu25, WY, and Rps1? Loci, Sattwd15-24, Sattwd15-25 and Sattwd15-47 for Rps10, SSR_07_0286, SSR_07_0300 for Rps11 and SSR_07_0295 for Rps1, Satt159 and SSR_03_0250 for RpsUN1, SSR_16_1275 and Sat144 for RpsUN2, and SattWM82-50 and Satt1k4b for RpsUD29 locus14,27,38,40 (Table 2). The 23 SSR markers were investigated for polymorphisms between the resistant PI399036 and susceptible AR2 parents. Of the 23 SSR markers, 10 SSR markers were polymorphic between the two parents (Fig. 1) and applied initially in evaluating all 60 RILs homozygous for Rps12; and subsequently, 60 Phytophthora susceptible RILs (rps12rps12). The ten polymorphic SSR markers considered for this study include Satt510 for Rps3 locus, Satt663 for Rps8, Satt440 for Rps2 and Satt631, Satt152 and Satt009 for Rps1, 7, 9, Yu25, WY, and Rps1?, Sattwd15-24 for Rps10, SSR_07_0286 for Rps11, and Satt159 and SSR_03_0250 for RpsUN1 (Table S3). From screening of the 60 resistant RILs, we identified RILs 12 and 14 that carry AR2-specific SSR alleles for nine and eight SSR markers, respectively. For RIL12, SSR marker linked to Rps11 is heterozygous; and for RIL14, two SSR markers linked to Rps8 and Rps11 are heterozygous. These two lines were selected to determine the efficacy of Rps12 to a set of P. sojae isolates.
Figure 1.
Polymorphisms between resistant (P) and susceptible (A) parents for the SSR markers linked to different Rps regions. P resistant parent PI399036; A susceptible parent AR2.
Identification of Rps13
We have obtained 17 P. sojae isolates from the Robertson lab, Iowa State University, collected earlier from the Iowa soybean fields. The isolates were characterized for their pathotypes by inoculating a set of 14 soybean lines that are considered to be differential lines for 14 individual Rps genes (Table 1). All these isolates were used to infect the differential cultivars and selected RIL12 and RIL14 and two parents, PI399036 and AR2. RIL12 and RIL14 contain Rps12 and confers resistance against the isolate mixture of R17 and Val 12-1117. Surprisingly, RIL12 is not resistant against seven of the 17 new P. sojae isolates and Val 12-11 (Table 1). On the contrary, RIL14 is resistant against these seven isolates. Based on the genetic make-ups of RIL12 and RIL14 for molecular markers of the Rps12 region, we deducted that there is recombination breakpoint in between the NBSLRR533 and Sat_064 in RIL12. We hypothesized that there could be a novel Rps gene named Rps13 located in between Rps12 and telomere on Chromosome 18.
To further support our hypothesis that there is an Rps gene next to Rps12, we evaluated 60 Phytopthora resistant RILs (Rps12Rps12) and 60 Phytopthora susceptible RILs (rps12rps12) for molecular markers of the genomic region containing the Rps12 gene (Table S4). From molecular mapping of the 120 RILs, we were able to identify two additional RILs, RIL9 and RIL81, that carry recombination breakpoints in the Rps12 region, and were evaluated for their responses to 17 new P. sojae isolates, and mixture of R17 and Val 12-11 isolates. The RIL12 contains Rps12, but not the putative Rps13 region; whereas, RIL81 contains the putative Rps13 region but not Rps12. RIL9 contains the putative Rps13 region, but not Rps12. RILs that carry Rps12, but not Rps13, were susceptible to the P. sojae isolates, V13, IV 6b and Val 12-11, resistant to R17 (Table 3). On the contrary, RIL81 carrying Rps13 but not Rps12 was susceptible to P. sojae isolate R17 (Table 3). Our results established that Rps12 is overcome by several P. sojae isolates, against which Rps13 provides immunity.
Table 3.
Response of four RILS and their parents to 23 P. sojae isolates along and a few P. sojae isolate mixtures.
| P. sojae isolate | Sloan | PI399036 (Rps12, Rps13) | AR2 (rps12, rps13) | RIL12 (Rps12, rps13) | RIL81 (rps12, Rps13) | RIL14 (Rps12, Rps13) | RIL9 (Rps12, rps13) |
|---|---|---|---|---|---|---|---|
| R17 (vir 1b, 1d, 3a, 3b, 3c, 5, 6) | S | R | S | R | S | R | R |
| Val 12-11 (vir 1a, 1b, 1c, 1d, 1k, 2, 4, 7) | S | R | S | R | R | R | R |
| 1005-2.9 (vir 1a, 1b, 1c, 1k, 3b, 7) | S | R | S | R | R | R | R |
| III 5.2b (vir 1a, 1b, 1c, 1d, 1k, 7) | S | R | S | R | R | R | R |
| III 23.4b (vir 1a, 1c, 1d, 2, 3b, 3c, 4, 5, 7) | S | R | S | R | R | R | S |
| IV 5.2 (vir 1c, 1d, 7) | S | R | R | R | R | R | R |
| IV 6b (vir 1a, 1c, 1d, 7) | S | R | S | S | R | R | S |
| IV 10 (vir 1a, 1c, 1d, 7) | S | R | S | R | R | R | R |
| IV 12.2a (vir 2, 4, 7) | S | R | S | R | R | R | R |
| IV 13.4a (vir 7) | S | R | S | R | R | R | R |
| IV 23.3 (vir 1a, 1c, 1d, 2, 7) | S | R | S | R | R | R | R |
| V 13 (vir 1a, 1c, 1d, 4, 7) | S | R | S | S | R | R | S |
| VI 5.2b (vir 1a, 1c, 1d, 7) | S | R | S | R | R | R | R |
| VI 12.1a (vir 6,7) | S | R | S | R | R | R | R |
| VI 12.2b (vir 1d, 3a, 4, 5, 6, 7) | S | R | S | S | S | R | S |
| VI 15(vir 1d, 7) | S | R | S | R | R | R | R |
| VI 17 (vir 7) | S | R | S | R | R | R | R |
| VI 20 (vir 1c, 2, 7) | S | R | S | S | R | R | S |
| VI 23.3b (vir 1d, 7) | S | R | S | R | R | R | R |
| S 5-5 (vir 7) | S | R | S | R | R | R | R |
| P7074 (vir 1b, 1d, 2, 3a, 3b, 3c, 4, 5, 6, 7, 8) | S | R | S | R | S | R | R |
| PR1 (vir 7) | S | R | S | R | R | R | R |
| PR6 (vir 7) | S | R | S | R | R | R | R |
| III5.2b + R17 + V13 (vir 1a, 1b, 1c, 1d, 1k, 3b, 5,7,8) | S | R | S | S | S | S | S |
| 1005–2.9 + VI23.3b + R17 (vir 1d, 2, 3b, 7) | S | R | S | R | R | R | R |
| R17 + Val 12–11 (vir 1a, 1b, 1c, 1d, 1k, 2, 3a, 3b, 3c, 4, 5, 6, 7) | S | R | S | R | S | R | R |
Plants were rated seven days after inoculation as either R (resistant, < 30% seedling death) or S (susceptible, ≥ 70% seedling death).
Molecular mapping of the Rps13 gene
We determined the inheritance of the putative novel Rps13 gene by evaluating 120 RILs for segregation of Phytophthora resistance against an inoculum mixture of Val 12-11 and R17, which together are virulent on soybean lines carrying all Phytophthora resistance genes mapped to the Rps1 to 7 loci and partially virulent to lines carrying Rps8 along with V13 isolate which is virulent to soybean lines carrying Rps1a, 1c, 1d, 4, 7 and Rps12 (Figs. 2, 3, Table 3).
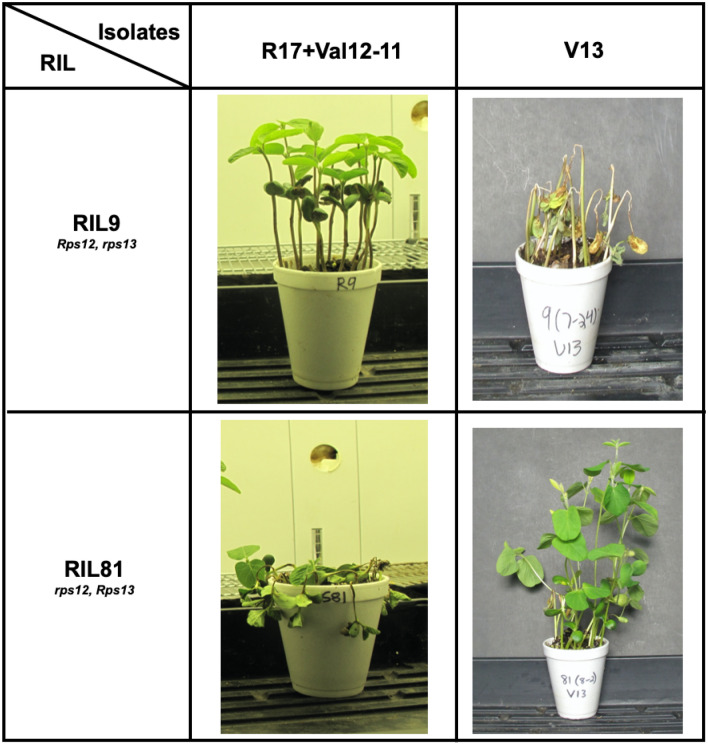
Figure 2.
Two RILs differing alleles at the linked Rps12 and Rps13 loci showed distinct responses to P. sojae V13 isolate and the mixture of R17 and Val12-11 isolates. P. sojae isolate V13 failed to defeat resistance mediated by Rps13 gene but could overcome that by Rps12; whereas, the mixture of R17 and Val12-11 isolates could overcome Rps13, but not Rps12.
Figure 3.
Phytophthora sojae isolate V13-specific resistance is conferred by Rps13. Reactions of susceptible parent AR2, resistant parent PI399036 containing Rps12 and Rps13 genes, recombinant inbred line RIL12 (6–14) susceptible to P. sojae isolate V13 due to absence of Rps13, recombinant inbred line RIL14 (1–10) resistant to P. sojae isolate V13 due to presence of Rps13 and the susceptible cultivar Sloan with no known Rps genes is susceptible to V13 isolate.
Analysis of Rps gene-linked SSR markers revealed that alleles of Satt009 and Satt510 markers specific to Rps1c and Rps3a alleles, respectively, are present in PI399036, but not in AR2. We hypothesized that most likely PI399036 contains Rps1c and Rps3a, in addition to Rps12 and Rps13. P. sojae isolate V13 overcomes the resistance conferred by Rps1c, but not Rps3a. We therefore classified the RILs into two groups based on Satt510: (i) The RILs which carry Satt510 allele specific to rps3a and AR2 parent; and (ii) RILs carry Satt510 allele specific to Rps3a. Both groups segregated for resistance to susceptibility in a 3:1 ratio, as observed for single Mendelian genes, following infection with P. sojae V13 isolate that overcomes the resistance governed by Rps12 and Rps1c. This confirms that there is a novel Rps gene in PI399036.
To map the novel gene, the 120 RILs from the AX20925 population were infected with a mixture of P. sojae R17 and V13 isolates that together overcome all known Rps genes including Rps12, but not the novel Rps13 gene. Of the 120 RILs, 52 RILs showed resistance against the isolate mix and 67 showed susceptibility. The observed segregating 0.867:0:0.08:1.117 genotypic ratio of resistance to susceptibility among the 120 RILs fits to the expected 0.984:0.032:0.984::RR:Rr:rr ratio, where R is Rps13 and r is rps13 for single gene segregation among the RILs in F7 generation with an estimated 98.4% of the genes homozygous (χ2 = 0.104).
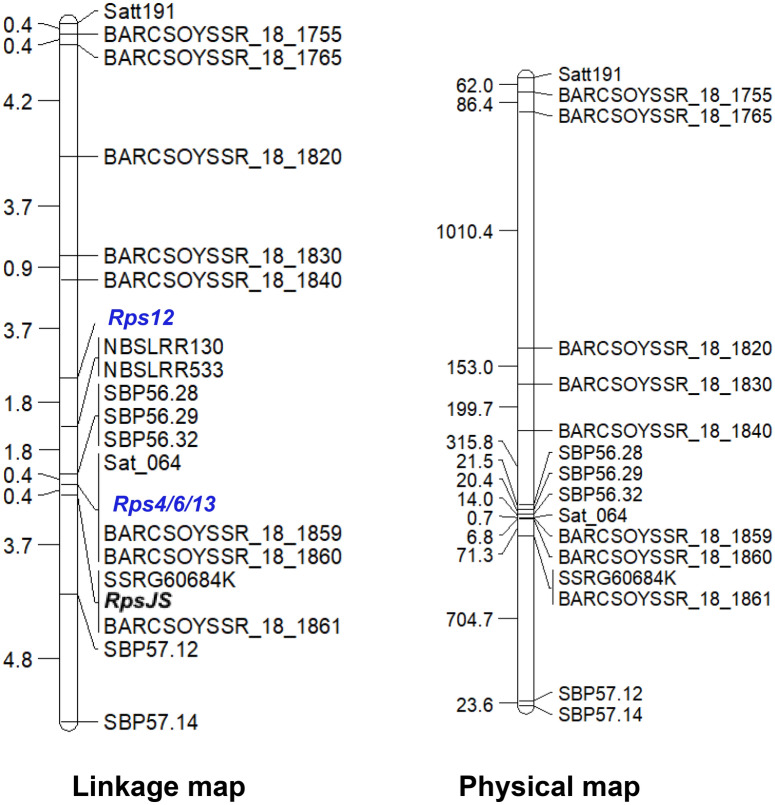
We conducted bulked segregant analysis (BSA) to identify molecular markers linked to the novel Rps13 resistance gene and confirm that Rps13 is mapped next to Rps1248. In this BSA study, we used SSR markers of the Rps12 region to test our hypothesis that Rps13 is linked to Rps12. The results of BSA suggested that indeed the gene is co-segregated with the markers mapped in between Rps12 and telomere. To develop a high-resolution map of the Rps13 region, we investigated 19 putative SBP markers for polymorphisms. Five of the 19 putative SBP markers are polymorphic between resistant and susceptible parents and were used for mapping the Rps12-Rps13 region (Fig. 4). The Rps13 gene co-segregated with the Sat_064, BARCOSOYSSR_18_1859, and BARCOSOYSSR_18_1860 markers. The genetic distance between Rps12 and Rps13 genes is 4 cM (Fig. 5, Table S4).
Figure 4.
Identification of sequence-based polymorphic (SBP) markers linked to Rps12 and Rps13 genes. AR2 susceptible parent AR2; PI resistant parent PI399036; Undigested (UD) and digested (as marked with respective restriction enzymes) PCR products for the SBP markers: SBP57.31, SBP56.59, SBP51.3, SBP55.611400, SBP57.21, SBP55.611380, SBP50.9, and SBP55.59. Primers and enzymes used for SBP markers are presented in Table S1.
Figure 5.
Linkage and physical map of the Rps4/6/12/13/JS region. (A) Genetic map of the Rps12-Rps13-RpsJS region. SSR and SBP markers are shown on the right side of the map and corresponding genetic distances between two adjacent loci in centi-Morgan (cM) on the left side of the map. Rps13 gene is mapped between Rps12 and RpsJS and tightly linked to Sat_064, BARCSOYSSR_18_1859 and BARCSOYSSR_18_1860 SSR markers. The placement of Rps4, 6, and JS on the map is based published work23,38. (B) The physical map positions of the SSR and SBP markers are based on the cultivar ‘Williams 82’ genome sequence (http://soybase.org). The physical distances between adjacent loci are presented in kilobases DNA (shown on the left side of the map).
To identify homologues of the candidate Rps13 genes, we investigated the annotated soybean genes in the 92.7 kb Rps13 region between the two markers, SBP56.32 and BARCSOYSSR_18_1861, in the Williams 82 genome sequence located at the soybean genome browser (SoyBase; https://www.soybase.org)53. Sixteen genes including an NB-ARC domain-containing disease resistance-like gene, Glyma.18g283200, are present in this region (Table S5). Williams 82 does not carry the Rps13 gene.
Discussion
This study was designed to investigate the usefulness of the Phytophthora resistance governed by the Rps12 gene17. In mapping Rps12, we had to use a mixture of P. sojae isolates to mask the effect of previously known Rps genes that were in the PI399036 line, the source of Rps1217. To determine the utility of Rps12 genes against a set of uncharacterized P. sojae isolates, we must identify an RIL that contains only Rps12. We therefor first examined a set of 60 Phytophthora resistant RILs for possible absence of other known Rps genes by studying the polymorphisms of Rps gene-linked SSR markers. Linked SSR markers co-evolved with linked Rps genes and SSR alleles can be used to predict alleles of the linked Rps genes.
A total of 210,990 SSRs were identified from the soybean genome. Of these, 61,458 SSRs contain repeat units of di-, tri-, and tetranucleotide with (AT)n, (ATT)n and (AAAT)n as the most abundant motifs54. A genetic linkage map consisting of 20 linkage groups with approximately 1500 SNP, 1000 SSR markers, 700 RFLP, and 73 RAPD markers and 46 classical trait loci is available in soybean55–57. Information of genetic markers has been used to map Rps1, Rps2, Rps3, Rps4, Rps5, Rps6, Rps7, and Rps8 loci to Chromosomes 3, 16, 13, 18 and 13, respectively24,25,31,32,35,57–59. While the Rps4 locus was mapped close to the Rps6 region, the Rps8 locus mapped close to the Rps3 region31,32,35. The RFLP marker pT-5 was shown to be linked to the Rps5 locus36. SSR markers mapped to the Rps5 locus are yet to be identified24. Thus, SSR markers linked to each Rps gene except Rps5 have been reported24.
In this study, we selected 23 SSR markers that have been shown to be linked to most of the reported Rps genes (Table 2). Out of 21 SSR markers, 10 were polymorphic between the two parents, PI399036 and AR2 (Fig. 1). These 10 SSR markers were applied in evaluating all 60 RILs homozygous for Rps12. These polymorphic markers included Satt510 linked to the Rps3 locus, Satt663 to Rps8, Satt440 to Rps2, and Satt631, Satt152 and Satt009 to Rps1, 7, 9, Yu25, WY, Rps1?, Sattwd15-24 to Rps10, SSR_07_0286 to Rps11, Satt159 and SSR_03_0250 to RpsUN1. PI399036, the source of Rps12, exhibitted the alleles of the Satt009 and Satt510 linked to linked to Rps1c and Rps3a alleles, respectively, suggesting that PI399036 most likely contains Rps1c and Rps3a genes as well.
From PCR assays of 60 Phytophthora resistant RILs with 10 SSR markers polymorphic between PI399036 and AR2, we identified RIL12 and RIL14 carrying rps alleles-specific SSR alleles for nine and eight SSR markers, respectively. For RIL12, SSR marker linked to Rps11 is heterozygous and for RIL14, two SSR markers linked to Rps8 and Rps11 are heterozygous. These two lines were selected to determine the efficacy of Rps12 to a set of 17 P. sojae isolates collected in Iowa soybean fields. RIL12 was susceptible to seven of the 17 P. sojae isolates; whereas, RIL14 was resistant to these seven isolates. Earlier both lines were shown to carry Rps1217. We hypothesize that RIL12 lacks an unknown Rps gene that is present in RIL14. In the absence of this unknown gene the RIL12 failed to provide immunity against four of the 17 isolates studied (Table 3). The putative unknown gene is named as Rps13. BSA revealed that the gene is linked to Rps12. Genetic mapping using 18 molecular markers placed the gene on the south arm of Chromosome 18, at a 4 cM genetic distance from Rps12. We observed that due to the absence of Rps13 in RILs 6, 9, 42, and 49 resulted in susceptibility to the P. sojae isolate V13. However, the four lines contain Rps12 and resistant to the mixture of the isolates, R17 and Val 12-11 that cannot overcome resistance encoded by Rps12. On the contrary, RIL81 contains Rps13 but not the Rps12 gene. Therefore, this RIL is resistant to V13 and susceptible to the mixture of R17 and Val 12-11 isolates (Fig. 2). These results established that there is a novel gene next to Rps12 that is essential for immunity of the RILs against four of the 17 P. sojae isolates collected in Iowa. Two linked functional Rps genes provide broad-spectrum resistance against P. sojae isolates tested in this study.
Plant activates defenses against pathogen attacks, determined by a corresponding pair of genes, a gene for avirulence in the pathogen and a gene for resistance (R) in the host. Such resistance mechanisms function in both major classes of flowering plants, dicots, and monocots. Clustering of R genes at a single locus is a well-reported, and many R genes are clustered in plant genomes, including soybean60, common bean61, Arabidopsis62–64, Brassicaceae62, wild potato65, tomato66,67, coffee trees68, wheat69 and rice70,71. The clustered distribution of R-genes provides a reservoir of genetic variation from which new pathogen specificity can evolve through gene duplication, ectopic recombination, unequal crossing-over and diversifying selection72. These clusters frequently comprise tandem arrays of genes that regulate resistance to multiple pathogens and to multiple variants of a single pathogen. The clusters may be tight with a little intervening sequence as 20 kb between two functional Rps1-k genes in soybean26,73, the RPP5 cluster in Arabidopsis thaliana spans 91 kb64, or be spread over several megabases as the Resistance Gene Candidate2 (RGC2) locus in lettuce (Lactuca sativa)74. In rice, also Chromosome 11 is highly enriched in R-genes, mostly in clusters; up to 201 loci encode the domains of NBS-LRR and LRR—receptor-like kinase (LRR-RLK) or wall-associated serine/threonine protein kinase (WAK)70.
The Rps12-Rps13 region is rich in Rps genes. As of now, Rps4, 6, 12, 13 and JS are mapped to the same genomic region spanning probably less than 5 cM in different soybean haplotypes (17,34,35 this work). Earlier we demonstrated that Rps4 and Rps6 are allelic and Rps4 co-segregates with Sat_06435. Therefore, most likely Rps13 is allele to Rps4 and Rps6. The PI399036, the donor of Rps12 and Rps13, does not carry Rps4 or Rps6 and therefore Rps13 is distinct from the two Rps genes17, this study.
The Rps13 locus is very close to the RpsJS locus (Fig. 5). Rps13 co-segregates with Sat_064, BARCSOYSSR_18_1859 and BARCSOYSSR_18_1860, and RpsJS co-segregates with SSRG60685K and BARCSOYSSR_18_1861. The genetic distance between BARCSOYSSR_18_1859 and BARCSOYSSR_18_1861 was reported to be 0.9 cM34. In our study, the genetic distance between these two SSR markers is 0.4 cM. The physical distance between BARCSOYSSR_18_1860 and BARCSOYSSR_18_1861 is 71 kb based on the soybean Williams 82 genome sequence (Fig. 5). The candidate annotated disease resistance gene-like sequence among the 10 predicted genes of the 92.7 kb Rps13 region between SBP56.32 and BARCSOYSSR_18_1861 markers in the Williams 82 genome is an NB-ARC domain-containing gene, Glyma.18g283200 (Table S5). There are three NB-LRR genes, Glyma18g51930, Glyma18g51950, and Glyma18g51960, identified from the RpsJS34 region between markers BARCSOYSSR_18_1859 and BARCSOYSSR_18_1861. The four NB-LRR genes with high similarity, are presumably paralogous sequences (Supplementary Fig. S1). They were identified from the Williams 82 haplotype that does not contain any known functional Rps genes. Based on the genetic and physical distances between BARCSOYSSR_18_1860 and BARCSOYSSR_18_1861 markers and differences in candidate NB-LRR-like resistance gene sequences, Rps13 and RpsJS are unlikely allelic or the same gene.
We propose that the five Rps genes, Rps4, 6, 12, 13 and JS, might have evolved from a single progenitor Rps gene. Identification of these Rps genes will shed light on how Rps genes evolved in soybean to confer effector triggered immunity against a serious oomycete pathogen, P. sojae.
In this study we have shown that the broad-spectrum Phytophthora resistance is encoded by two Rps genes, Rps12 and Rps13, with distinct race-specificity. The genetic distance between the two Rps genes is 4 cM. Therefore, to maintain the broad-spectrum Phytophthora resistance encoded by Rps12 and Rps13, we must select both genes using molecular markers. We report here several SSR markers that should be ideal for introgressing Rps12 and Rps13 into new soybean cultivars.
Supplementary Information
Acknowledgements
We are thankful to Drs. Anne Dorrance, Martin Chilvers, and Alison E Robertson, who kindly provided us the P. sojae isolates used in this study. We are thankful to Iowa Soybean Association for the funding support.
Author contributions
M.K.B. conceived the project and received the grant to conduct the research. D.K.S. conducted all biological experiments. A.D. and X.H. conducted soybean genome analyses for identifying the polymorphic nucleotides of the Rps12/Rps13 genomic region. D.K.S. prepared all figures and tables and wrote the first draft of the manuscript. M.K.B. supervised D.K.S. and prepared the final draft. All authors reviewed the manuscript.
Funding
This work was funded by Iowa Soybean Association.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-96425-1.
References
- 1.Savary S, et al. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019;3:430–439. doi: 10.1038/s41559-018-0793-y. [DOI] [PubMed] [Google Scholar]
- 2.Hartman GL, et al. Compendium of Soybean Diseases and Pests. 5. The American Phytopathological Society; 2016. [Google Scholar]
- 3.Allen TW, et al. Soybean yield loss estimates due to diseases in the United States and Ontario, Canada, from 2010 to 2014. Plant Health Prog. 2017;18:19–27. doi: 10.1094/PHP-RS-16-0066. [DOI] [Google Scholar]
- 4.Wrather J, Koenning S. Effects of diseases on soybean yields in the United States 1996 to 2007. Plant Health Prog. 2009;10:24. doi: 10.1094/PHP-2009-0401-01-RS. [DOI] [Google Scholar]
- 5.Kaufmann M, Gerdemann J. Root and stem rot of soybean caused by Phytophthora sojae n sp. Phytopathology. 1958;48:201–208. [Google Scholar]
- 6.Sugimoto T, et al. Pathogenic diversity of Phytophthora sojae and breeding strategies to develop Phytophthora -resistant soybeans. Breed. Sci. 2012;61:511–522. doi: 10.1270/jsbbs.61.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chamnanpunt J, Shan WX, Tyler BM. High frequency mitotic gene conversion in genetic hybrids of the oomycete Phytophthora sojae. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14530–14535. doi: 10.1073/pnas.251464498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmitthenner A. Problems and progress in control of Phytophthora root-rot of soybean. Plant Dis. 1985;69:362–368. doi: 10.1094/PD-69-362. [DOI] [Google Scholar]
- 9.Anderson TR. Efficacy of metalaxyl in controlling phytophthora root and stalk rot of soybean cultivars differing in field tolerance. Plant Dis. 1982;66:1144–1145. doi: 10.1094/PD-66-1144. [DOI] [Google Scholar]
- 10.Dorrance A, McClure S, de Silva A. Pathogenic diversity of Phytophthora sojae in Ohio soybean fields. Plant Dis. 2003;87:139–146. doi: 10.1094/PDIS.2003.87.2.139. [DOI] [PubMed] [Google Scholar]
- 11.Schmitthenner AF. Phytophthora rot of soybean. Plant Health Prog. 2000;1:13. doi: 10.1094/PHP-2000-0601-01-HM. [DOI] [Google Scholar]
- 12.Workneh F, Yang XB, Tylka GL. Effect of tillage practices on vertical distribution of Phytophthora sojae. Plant Dis. 1998;82:1258–1263. doi: 10.1094/PDIS.1998.82.11.1258. [DOI] [PubMed] [Google Scholar]
- 13.Sugimoto T, et al. Pathogenic diversity of Phytophthora sojae and breeding strategies to develop Phytophthora-resistant soybeans. Breed. Sci. 2012;61:511–522. doi: 10.1270/jsbbs.61.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sugimoto T, et al. Genetic analysis and identification of DNA markers linked to a novel Phytophthora sojae resistance gene in the Japanese soybean cultivar Waseshiroge. Euphytica. 2011;182:133–145. doi: 10.1007/s10681-011-0525-8. [DOI] [Google Scholar]
- 15.Lamour K, Kamoun S. Oomycete Genetics and Genomics: Diversity, Interactions and Research Tools. Wiley; 2009. [Google Scholar]
- 16.Dong S, et al. Sequence variants of the Phytophthora sojae RXLR effector Avr3a/5 are differentially recognized by Rps3a and Rps5 in soybean. PLoS ONE. 2011;6:e20172. doi: 10.1371/journal.pone.0020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sahoo DK, Abeysekara NS, Cianzio SR, Robertson AE, Bhattacharyya MK. A novel Phytophthora sojae resistance Rps12 gene mapped to a genomic region that contains several Rps genes. PLoS ONE. 2017;12:e0169950. doi: 10.1371/journal.pone.0169950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van K, et al. Mining germplasm panels and phenotypic datasets to identify loci for resistance to Phytophthora sojae in soybean. Plant Genome. 2020 doi: 10.1002/tpg2.20063. [DOI] [PubMed] [Google Scholar]
- 19.Bernard R, Smith P, Kaufmann M, Schmitthenner A. Inheritance of resistance to Phytophthora root and stem rot in the soybean. Agron. J. 1957;49:391. doi: 10.2134/agronj1957.00021962004900070016x. [DOI] [Google Scholar]
- 20.Buzzell R, Anderson T. Inheritance and race reaction of a new soybean Rps1 allele. Plant Dis. 1992;76:600–601. doi: 10.1094/PD-76-0600. [DOI] [Google Scholar]
- 21.Mueller H, Athow KL, Laviolette FA. Genetics inheritance of resistance to four physiologic races of Phytophthora megasperma var. sojae. Phytopathology. 1978;68:1318–1322. doi: 10.1094/Phyto-68-1318. [DOI] [Google Scholar]
- 22.Ploper LD, Athow KL, Laviolette FA. A new allele at Rps3 locus for resistance to Phytophthora megasperma f. sp. glycinea in soybean. Phytopathology. 1985;75:690–694. doi: 10.1094/Phyto-75-690. [DOI] [Google Scholar]
- 23.Zhang J, et al. Genetic characterization and fine mapping of the novel Phytophthora resistance gene in a Chinese soybean cultivar. Theor. Appl. Genet. 2013;126:1555–1561. doi: 10.1007/s00122-013-2073-1. [DOI] [PubMed] [Google Scholar]
- 24.Demirbas A, et al. Simple sequence repeat markers linked to the soybean Rps genes for Phytophthora resistance. Crop Sci. 2001;41:1220–1227. doi: 10.2135/cropsci2001.4141220x. [DOI] [Google Scholar]
- 25.Weng C, Yu K, Anderson TR, Poysa V. Mapping genes conferring resistance to Phytophthora root rot of soybean, Rps1a and Rps7. J. Hered. 2001;92:442–446. doi: 10.1093/jhered/92.5.442. [DOI] [PubMed] [Google Scholar]
- 26.Gao H, Narayanan N, Ellison L, Bhattacharyya M. Two classes of highly similar coiled coil-nucleotide binding-leucine rich repeat genes isolated from the Rps1-k locus encode Phytophthora resistance in soybean. Mol. Plant Microbe Interact. 2005;18:1035–1045. doi: 10.1094/MPMI-18-1035. [DOI] [PubMed] [Google Scholar]
- 27.Sun S, et al. Characterization and mapping of RpsYu25, a novel resistance gene to Phytophthora sojae. Plant Breed. 2011;130:139–143. doi: 10.1111/j.1439-0523.2010.01794.x. [DOI] [Google Scholar]
- 28.Wu XL, et al. Identification, genetic analysis and mapping of resistance to Phytophthora sojae of Pm28 in soybean. Agric. Sci. China. 2011;10:1506–1511. doi: 10.1016/S1671-2927(11)60145-4. [DOI] [Google Scholar]
- 29.Lin F, et al. Molecular mapping of two genes conferring resistance to Phytophthora sojae in a soybean landrace PI 567139B. Theor. Appl. Genet. 2013;126:2177–2185. doi: 10.1007/s00122-013-2127-4. [DOI] [PubMed] [Google Scholar]
- 30.Kilen TC, Hartwig EE, Keeling BL. Inheritance of a second major gene for resistance to phytophthora rot in soybeans 1. Crop Sci. 1974;14:260–262. doi: 10.2135/cropsci1974.0011183X001400020027x. [DOI] [Google Scholar]
- 31.Gordon SG, St. Martin SK, Dorrance AE. Rps8 maps to a resistance gene rich region on soybean molecular linkage group F. Crop Sci. 2006;46:168–173. doi: 10.2135/cropsci2004.04-0024. [DOI] [Google Scholar]
- 32.Sandhu D, et al. Soybean Phytophthora resistance gene Rps8 maps closely to the Rps3 region. J. Hered. 2005;96:536–541. doi: 10.1093/jhered/esi081. [DOI] [PubMed] [Google Scholar]
- 33.Yu A, et al. Genetic analysis and SSR mapping of gene resistance to Phytophthora sojae race 1 in soybean cv Suinong 10. Chin. J. Oil Crop Sci. 2010;32:462–466. [Google Scholar]
- 34.Sun J, et al. Genetic analysis and fine mapping of RpsJS, a novel resistance gene to Phytophthora sojae in soybean [Glycine max (L.) Merr.] Theor. Appl. Genet. 2014;127:913–919. doi: 10.1007/s00122-014-2266-2. [DOI] [PubMed] [Google Scholar]
- 35.Sandhu D, Gao H, Cianzio S, Bhattacharyya MK. Deletion of a disease resistance nucleotide-binding-site leucine-rich-repeat-like sequence is associated with the loss of the Phytophthora resistance gene Rps4 in soybean. Genetics. 2004;168:2157–2167. doi: 10.1534/genetics.104.032037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diers BW, Mansur L, Imsande J, Shoemaker RC. Mapping phytophthora resistance loci in soybean with restriction fragment length polymorphism markers. Crop Sci. 1992;32:377–383. doi: 10.2135/cropsci1992.0011183X003200020020x. [DOI] [Google Scholar]
- 37.Yao HY, Wang XM, Wu XF, Xiao YN, Zhu ZD. Molecular mapping of Phytophthora resistance gene in soybean cultivar zaoshu18. J. Plant Genet. Resour. 2010;11:213–217. [Google Scholar]
- 38.Ping J, et al. Identification and molecular mapping of Rps11, a novel gene conferring resistance to Phytophthora sojae in soybean. Theor. Appl. Genet. 2016;129:445–451. doi: 10.1007/s00122-015-2638-2. [DOI] [PubMed] [Google Scholar]
- 39.Yan H, Nelson B. Adaptation of Phytophthora sojae to Rps resistance genes over the past two decades in North Dakota. Plant Health Prog. 2019;20:88–93. doi: 10.1094/PHP-10-18-0062-RS. [DOI] [Google Scholar]
- 40.Gordon SG, Berry SA, St. Martin SK, Dorrance AE. Genetic analysis of soybean plant introductions with resistance to Phytophthora sojae. Phytopathology. 2007;97:106–112. doi: 10.1094/PHYTO-97-0106. [DOI] [PubMed] [Google Scholar]
- 41.Abeysekara NS, Matthiesen RL, Cianzio SR, Bhattacharyya MK, Robertson AE. Novel sources of partial resistance against Phytophthora sojae in soybean PI 399036. Crop Sci. 2016;56:2322–2335. doi: 10.2135/cropsci2015.09.0578. [DOI] [Google Scholar]
- 42.Dorrance A, Berry S, Anderson T, Meharg C. Isolation, storage, pathotype characterization, and evaluation of resistance for Phytophthora sojae in soybean. Plant Health Prog. 2008;9:35. doi: 10.1094/PHP-2008-0118-01-DG. [DOI] [Google Scholar]
- 43.Stewart S, Abeysekara N, Robertson A. Pathotype and genetic shifts in a population of Phytophthora sojae under soybean cultivar rotation. Plant Dis. 2014;98:614–624. doi: 10.1094/PDIS-05-13-0575-RE. [DOI] [PubMed] [Google Scholar]
- 44.Dorrance AE, Robertson AE, Cianzo S. Integrated management strategies for Phytophthora sojae combining host resistance and seed treatments. Plant Dis. 2009;93:875–882. doi: 10.1094/PDIS-93-9-0875. [DOI] [PubMed] [Google Scholar]
- 45.Matthiesen RL, et al. A method for combining isolates of Phytophthora sojae to screen for novel sources of resistance to phytophthora stem and root rot in soybean. Plant Dis. 2016;100:1424–1428. doi: 10.1094/PDIS-08-15-0916-RE. [DOI] [PubMed] [Google Scholar]
- 46.Dorrance AE, Jia H, Abney TS. Evaluation of soybean differentials for their interaction with Phytophthora sojae. Plant Health Prog. 2004;5:9. doi: 10.1094/PHP-2004-0309-01-RS. [DOI] [Google Scholar]
- 47.Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 2006;1:2320–2325. doi: 10.1038/nprot.2006.384. [DOI] [PubMed] [Google Scholar]
- 48.Michelmore RW, Paran I, Kesseli RV. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. U.S.A. 1991;88:9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sahu BB, Sumit R, Srivastava SK, Bhattacharyya MK. Sequence based polymorphic (SBP) marker technology for targeted genomic regions: Its application in generating a molecular map of the Arabidopsis thaliana genome. BMC Genomics. 2012;13:20. doi: 10.1186/1471-2164-13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lander ES, et al. MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 51.Kosambi DD. The estimation of map distances from recombination values. Ann. Eugen. 1943;12:172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x. [DOI] [Google Scholar]
- 52.Voorrips RE. Mapchart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 53.SoyBase.org. Accessed 16 Feb 2017. https://www.soybase.org/.
- 54.Song Q, et al. Abundance of SSR motifs and development of candidate polymorphic SSR markers (BARCSOYSSR_1.0) in Soybean. Crop Sci. 2010;50:1950–1960. doi: 10.2135/cropsci2009.10.0607. [DOI] [Google Scholar]
- 55.Hyten DL, et al. A high density integrated genetic linkage map of soybean and the development of a 1536 universal soy linkage panel for quantitative trait locus mapping. Crop Sci. 2010;50:960–968. doi: 10.2135/cropsci2009.06.0360. [DOI] [Google Scholar]
- 56.Song QJ, et al. A new integrated genetic linkage map of the soybean. Theor. Appl. Genet. 2004;109:122–128. doi: 10.1007/s00122-004-1602-3. [DOI] [PubMed] [Google Scholar]
- 57.Mapped Soybean SSR Loci July 2003: Soybean Genomics and Improvement Laboratory (SGIL), Beltsville Agricultural Research Center, USDA. Accessed 14 Mar 2017. https://sgil.ba.ars.usda.gov/cregan/soy_map1.html.
- 58.Cregan PB, et al. An integrated genetic linkage map of the soybean genome. Crop Sci. 1999;39(5):1464–1490. doi: 10.2135/cropsci1999.3951464x. [DOI] [Google Scholar]
- 59.Lohnes DG, Schmitthenner AF. Position of the phytophthora resistance gene Rps7 on the soybean molecular map. Crop Sci. 1997;37:555–556. doi: 10.2135/cropsci1997.0011183X003700020040x. [DOI] [Google Scholar]
- 60.Innes RW, et al. Genome analysis differential accumulation of retroelements and diversification of NB-LRR disease resistance genes in duplicated regions following polyploidy in the ancestor of soybean 1[W][OA] Plant Physiol. 2008;148:1740–1759. doi: 10.1104/pp.108.127902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.David P, et al. A Nomadic subtelomeric disease resistance gene cluster in common bean. Plant Physiol. 2009;151:1048–1065. doi: 10.1104/pp.109.142109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiao S. Origin and maintenance of a broad-spectrum disease resistance locus in Arabidopsis. Mol. Biol. Evol. 2004;21:1661–1672. doi: 10.1093/molbev/msh165. [DOI] [PubMed] [Google Scholar]
- 63.Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Noël L, et al. Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell. 1999;11:2099–2111. doi: 10.1105/tpc.11.11.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Destefanis M, et al. A disease resistance locus on potato and tomato chromosome 4 exhibits a conserved multipartite structure displaying different rates of evolution in different lineages. BMC Plant Biol. 2015;15:1–13. doi: 10.1186/s12870-015-0645-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kruijt M, Brandwagt BF, De Wit PJ. Rearrangements in the Cf-9 disease resistance gene cluster of wild tomato have resulted in three genes that mediate Avr9 responsiveness. Genetics. 2004;168:1655–1663. doi: 10.1534/genetics.104.028985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seah S, Telleen AC, Williamson VM. Introgressed and endogenous Mi-1 gene clusters in tomato diVer by complex rearrangements in Xanking sequences and show sequence exchange and diversifying selection among homologues. Theor. Appl. Genet. 2007;114:1289–1302. doi: 10.1007/s00122-007-0519-z. [DOI] [PubMed] [Google Scholar]
- 68.Ribas AF, Cenci A, Combes MC, Etienne H, Lashermes P. Organization and molecular evolution of a disease-resistance gene cluster in coffee trees. BMC Genomics. 2011;12:1–12. doi: 10.1186/1471-2164-12-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wicker T, Yahiaoui N, Keller B. Contrasting rates of evolution in Pm3 loci from three wheat species and rice. Genetics. 2007;177:1207–1216. doi: 10.1534/genetics.107.077354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mizuno H, et al. Evolutionary dynamics and impacts of chromosome regions carrying R-gene clusters in rice. Sci. Rep. 2020;10:1–9. doi: 10.1038/s41598-019-56847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dai L, et al. Genomic structure and evolution of the Pi2/9 locus in wild rice species. Theor. Appl. Genet. 2010;121:295–309. doi: 10.1007/s00122-010-1310-0. [DOI] [PubMed] [Google Scholar]
- 72.Michelmore RW, Meyers BC. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 1998;8:1113–1130. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- 73.Gao H, Bhattacharyya MK. The soybean-Phytophthora resistance locus Rps1-k encompasses coiled coil-nucleotide binding-leucine rich repeat-like genes and repetitive sequences. BMC Plant Biol. 2008;8:29. doi: 10.1186/1471-2229-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Meyers BC, et al. The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell. 1998;10:1817–1832. doi: 10.1105/tpc.10.11.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cheng Y, et al. Fine mapping of a Phytophthora-resistance gene RpsWY in soybean (Glycine max L.) by highthroughput genome-wide sequencing. Theor. Appl. Genet. 2017;130(5):1041–1051. doi: 10.1007/s00122-017-2869-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.