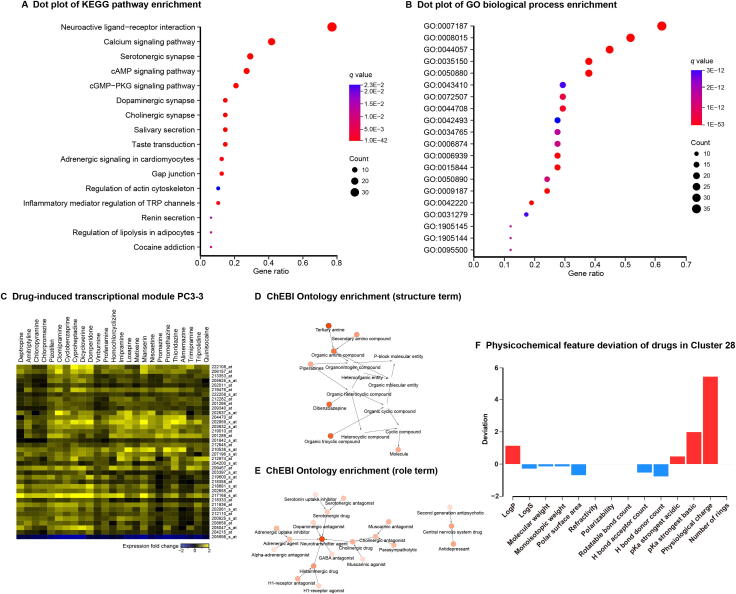
Figure 6.
Community analysis of Cluster 28
A. Dot plot of KEGG pathway enrichment results. B. Dot plot of GO biological process enrichment results. Dot size and color indicate the count of enriched genes in each of the categories and the corresponding significance of enrichment, respectively. Gene ratio represents the ratio of enriched genes to all genes in each of the categories. GO IDs are presented here for simplicity. The list of corresponding GO terms associated with these IDs can be found in Table S9. C. Drug-induced transcriptional module PC3-3. The transcriptional module data were obtained from the study of Iskar and colleagues [69]. Drug enrichment analysis was performed using CGEA. The horizontal axis represents the drug name and the vertical axis represents the gene probe ID, respectively. D. ChEBI Ontology structure term enrichment results. E. ChEBI Ontology role term enrichment results. Nodes indicate the enriched ChEBI terms. The lower the node transparency is, the more significantly the term is enriched. Arrows proceed from child to parent terms. F. Physicochemical feature deviation of drugs in Cluster 28. Red and blue bars represent the indicated drug feature of Cluster 28 that is higher or lower than the average of all drugs in the DrugBank database, respectively. CGEA, chemogenomic enrichment analysis; ChEBI, Chemical Entities of Biological Interest.