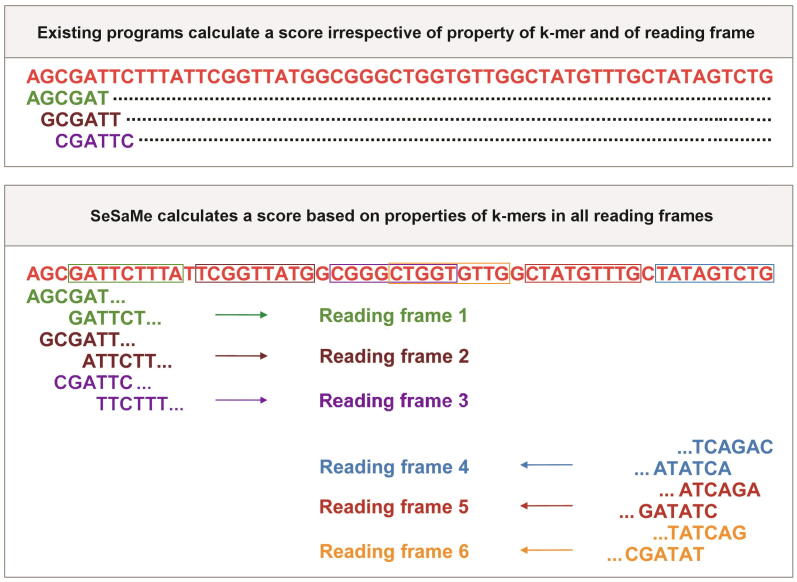
Figure 1.
Unique advantage of SeSaMe over existing programs
Existing programs calculate a score based on the frequencies of k-mers identified in a query sequence irrespective of properties of the k-mers, or their reading frames. In contrast, SeSaMe identifies k-mers that encode the amino acids of protein secondary structures in each reading frame. In the figure, matching 3-codon DNA 9-mers of the Trimer Ref. DB are marked with rectangles, where a color of the rectangle indicates a reading frame. SeSaMe calculates scores based on the 3-codon usages and the A.A. Trimer usages of the matching 3-codon DNA 9-mers in each reading frame. It classifies a query sequence into a taxonomic group based on the six scores computed from all reading frames. All sequences in this figure are randomly generated for illustration purposes only. SeSaMe, Spore-associated Symbiotic Microbes; A.A., amino acid; Trimer Ref. DB, trimer reference sequence database.