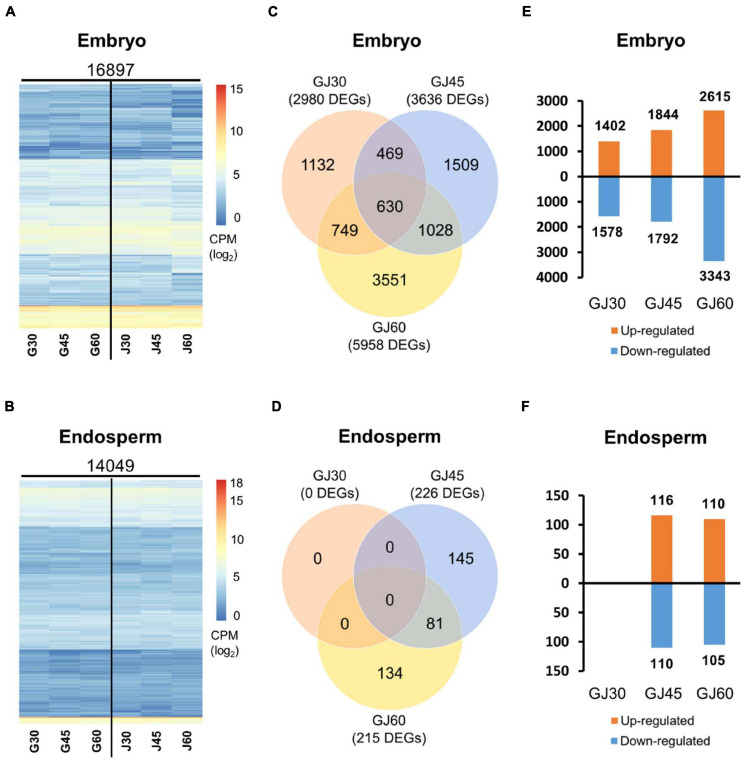
FIGURE 2.
Identification of differentially expressed genes (DEGs) based on the RNA-seq data of the embryo and endosperm of Gopum and Jowoon. (A,B) Heatmaps of expressed genes in the embryo (A) and endosperm (B) of Gopum and Jowoon, respectively. Expressed genes, mapped onto the International Rice Genome Sequencing Project (IRGSP) rice reference genome sequence, are shown as log-transformed counts per million (CPM) values. G30, G45, G60, J30, J45, and J60 indicate Gopum and Jowoon samples at 30, 45, and 60 DAH, respectively. (C,D) Comparison of Gopum and Jowoon based on the number of DEGs in the embryo (C) and endosperm (D). GJ30, GJ45, and GJ60 indicate the comparisons between Gopum and Jowoon at 30, 45, and 60 DAH, respectively. (E,F) Numbers of genes up- or down-regulated in Jowoon compared with those in Gopum in the embryo (E) and endosperm (F) of GJ30, GJ45, and GJ60.