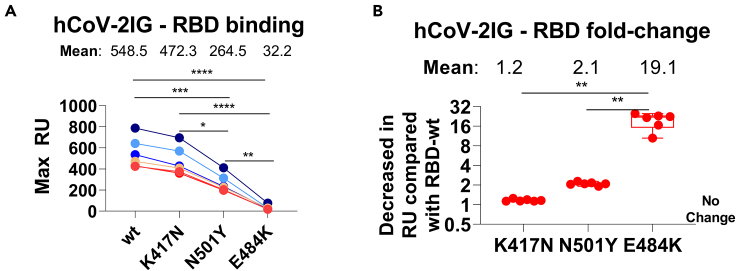
Figure 3.
RBD binding antibodies of convalescent plasma and hCoV-2IG against various SARS-CoV-2 RBD mutant proteins
(A and B) Total antibody binding (Max RU) of 1mg/mL for the six batches of hCoV-2IG (hCoV-2IG-1 to hCoV-2IG-6) to purified WA-1 RBD (RBD-wt) and RBD mutants: RBD-K417N, RBD-N501Y and RBD-E484K by SPR (A). The numbers above the group show the mean antibody binding for each RBD. (B) The fold-decrease in antibody binding to mutants RBD-K417N, RBD-N501Y and RBD-E484K of hCoV-2IG in comparison with RBD-wt from WA-1 strain was calculated from the data in Panel A. The numbers above the group shows the mean fold-change for each mutant RBD. All SPR experiments were performed twice and the researchers performing the assay were blinded to sample identity. The variations for duplicate runs of SPR was <5%. The data shown are average values of two experimental runs. The statistical significances between the variants for hCoV-2IG were performed using One-way ANOVA using Tukey's pairwise multiple comparison test in GraphPad prism. The differences were considered statistically significant with a 95% confidence interval when the p value was less than 0.05. (∗, p values of ≤0.05, ∗∗, p values of ≤0.01, ∗∗∗, p values of ≤0.001, ∗∗∗∗, p ≤ 0.0001).