Fig. 2. PhoP-mediated ATP reduction stabilizes protease substrates under Mg2+ starvation.
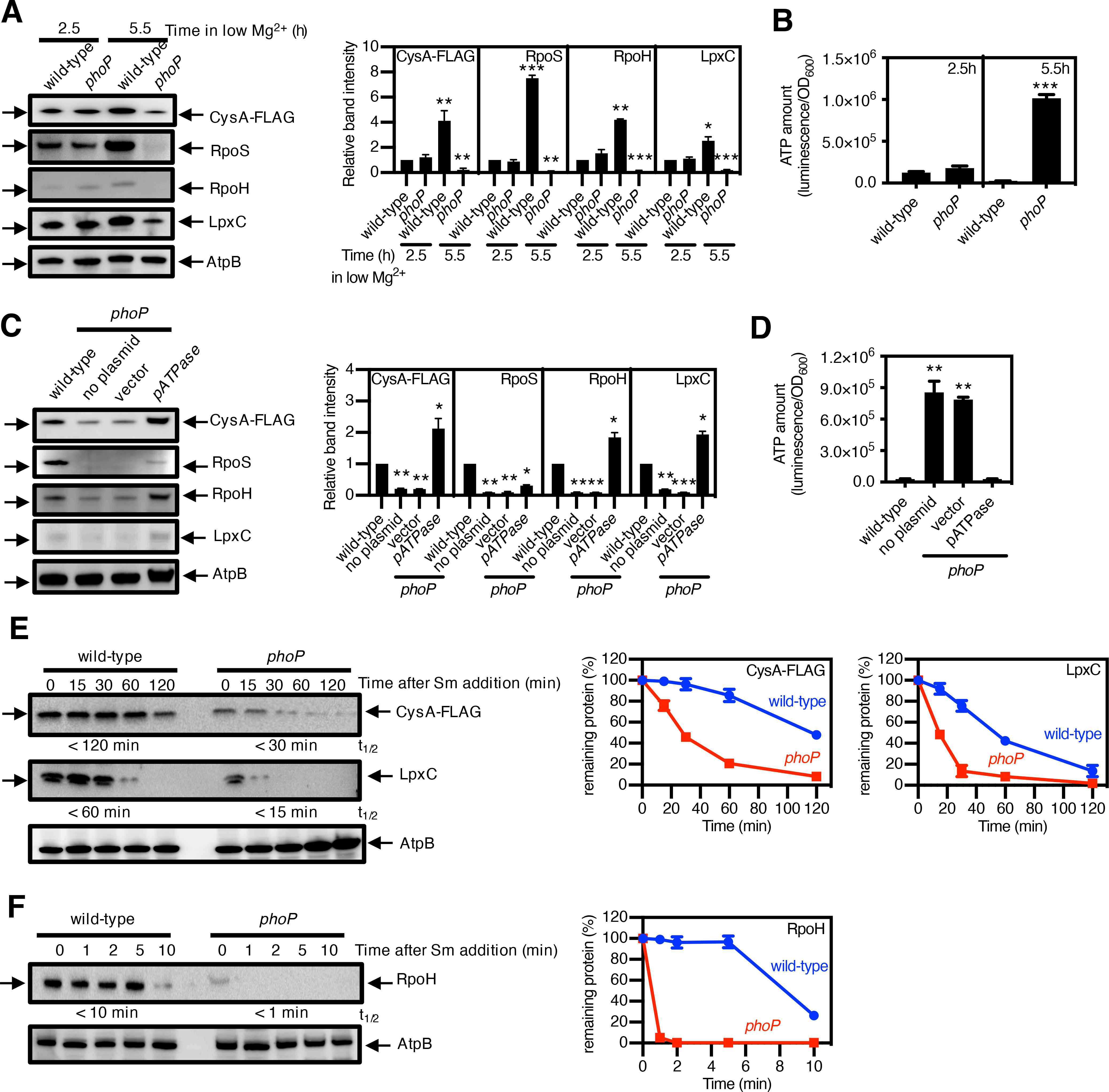
(A) Immunoblotting and quantification of the indicated protease substrates and (B) quantification of ATP amount in wild-type S. Typhimurium expressing cysA-FLAG (JY767, wild-type) and phoP S. Typhimurium expressing cysA-FLAG (JY901, phoP) grown under low-Mg2+ conditions for 2.5 or 5.5 h. AtpB is a loading control. (C) Immunoblotting and quantification of the indicated protease substrates and (D) quantification of ATP amount in strain JY767 (wild-type) and in strain JY901 with no plasmid, the plasmid vector alone (45), or expressing the soluble subunit of the F1Fo ATPase (pATPase) (54), grown under low-Mg2+ conditions for 5.5 h. (E and F) Immunoblotting and degradation curves of the indicated protease substrates in strains JY767 (wild-type) and JY901 (phoP) grown under low-Mg2+ conditions for 5.5 h with the protein synthesis inhibitor spectinomycin. Numbers below blots correspond to the half-lives (t1/2) of the indicated substrates, which were calculated by regression analysis of the exponential decay of the proteins. Western blotting (A, C, E, F) was performed with antibodies directed to the FLAG peptide or to the AtpB, RpoS, RpoH, or LpxC proteins, and degradation curves show the mean and SD for protein amounts normalized to AtpB from 3 independent experiments. ATP amounts (B and D) were calculated with normalization to luminescence by OD600 and represent the mean and SD from 3 independent experiments. Unpaired Student’s t tests were performed between wild-type and the mutant strains; ***P < 0.001, **P < 0.01.
