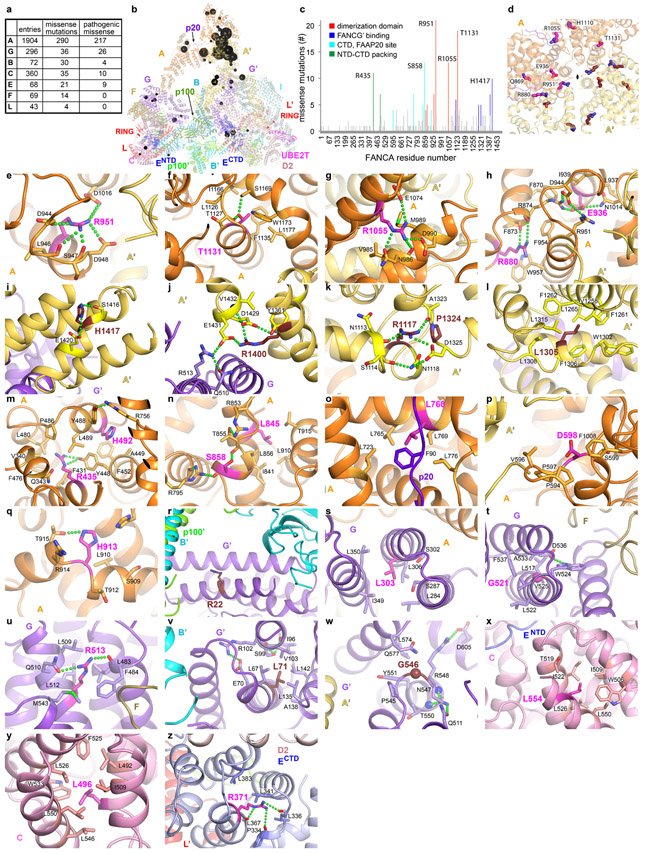
Extended Data Fig. 8. Fanconi Anemia missense mutations in the Core complex.
a, Summary of mutations in Core complex subunits from the Fanconi Anemia Mutation Database (http://www2.rockefeller.edu/fanconi/). For each subunit, the total number of database entries, the subset that represents missense mutations, and the missense mutations annotated as pathogenic or likely pathogenic are listed. The overall number of missense mutations is much smaller than other types of alterations. The most prevalent alterations are intragenic deletions in FANCA, suggested to correlate with the number of intronic Alu repeats, as well as microdeletions and insertions at homopolymeric tracts or direct repeats48,49.
b, The pathogenic missense mutations are mapped onto the Core-ID-DNA structure, rendered semi-transparent, in an orientation as in Figure 1a. Residues reported mutated two or more times are shown as spheres with a diameter proportional to the number of mutations in the database, except for those with 2 to 4 mutations, which have the same, smallest diameter.
c, Column graph of the number of pathogenic missense mutations in the FANCA protein, with the frequently mutated residues colored by their vicinity to the various structural and functional elements of the protein as indicated. Residues mutated 10 or more times are labeled.
d, Overall view of the mutated residues (thick sticks) in the vicinity of the FANCA homodimer interface (semi-transparent cartoon), looking down the approximate 2-fold axis indicated by a meniscus. Inactive-side FANCA mutants are in magenta (labeled), and those of the active-side FANCA’ in ruby.
e-h, Close-up views of mutations from d. The mutations are discussed in Supplementary Note 5.
i-l, Close-up views of FANCA mutations near the FANCA’ CTD that packs with FANCG’.
m-q, Close-up views of other FANCA mutations.
r-w, Close-up views of FANCG mutations.
x-y, Close-up views of FANCC mutations.
z, Close-up views of a FANCE mutation.