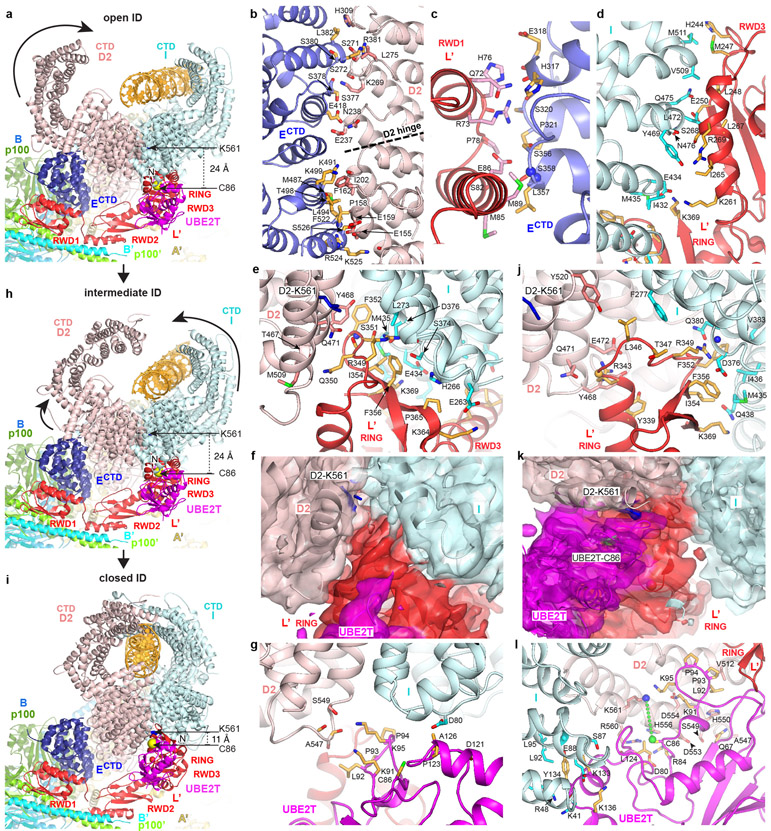
Figure 3 ∣. Core-UBE2T binding to ID in three conformations.
a, View focusing on FANCECTD-FANCL’-UBE2T partway encircling the ID complex in the open-conformation, looking down the end of the trough-like shape that has the FANCI N-terminus (labeled). Curved arrow indicates the FANCD2 CTD rotation in the transition to the intermediate conformation of h. Colored as in Figure 1c. Lys561 (blue sticks) and UBE2T Cys86 (yellow sphere) are labeled. b, FANCD2-FANCECTD interface showing side chain and backbone groups within interaction distance (3.8 Å). Dashed line indicates the hinge where the FANCD2 N-terminal segment rotates away from FANCI. c-e, Open-conformation interfaces between FANCL’ RWD1 and FANCECTD (c), FANCL’ RWD3 and FANCI (d), and FANCL’ RING and the ID complex (e). f, Open-conformation cryo-EM density (semi-transparent surface) at 4.2 Å of the RING-ID interface in an orientation similar to e and colored according to the underlying protein. Portions of ID and most of the UBE2T (magenta) are clipped for clarity. g, Open-conformation UBE2T-ID interface. h, View of the intermediate ID conformation in the same Core orientation as in a. Curved arrows indicate the overall rotations in FANCI and FANCD2 (excluding residues 1-160) in the transition to the closed conformation of i. i, View of the closed ID conformation in the same Core orientation as in a. j, Closed-conformation FANCL’ RING-ID interface. k, Closed-conformation cryo-EM density (semi-transparent surface) at 4.2 Å of the interface between ID and UBE2T-FANCL’ RING, colored according to the underlying protein. Portions of ID uninvolved in contacts are clipped for clarity. l, Closed-conformation UBE2T-ID interface.