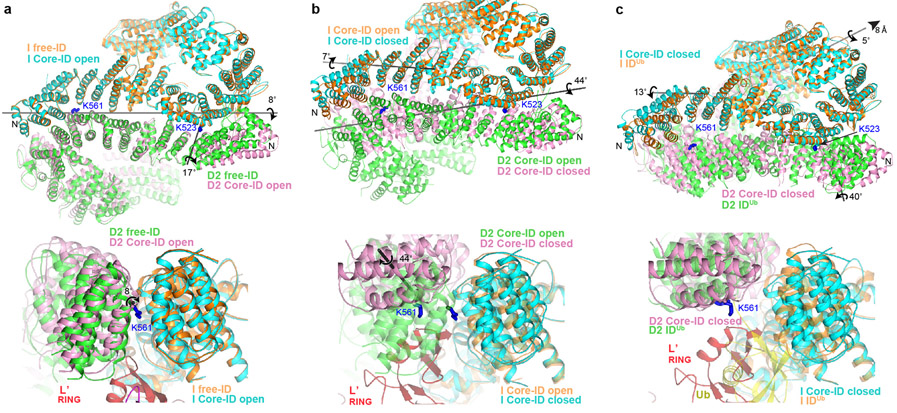
Figure 4 ∣. Core-UBE2T remodels the ID complex prior to ubiquitination.
a, Superposition of the Core-bound open-conformation ID and free ID aligned on their FANCI NTD domains, colored as marked. Top, overall view looking down the exterior bottom of the trough-like ID. The horizontal gray line indicates the axis of rotation about which the ID NTD-NTD interface is pried apart on initial Core binding, while the shorter near-vertical line marks the rotation axis of the FANCD2 N-terminal segment. The ubiquitination site lysine side chains (labeled) are shown as blue sticks. Bottom, close-up of superposition viewed down the horizontal rotation axis, marked with circular arrow, also showing the FANCL’ RING hairpin that cannot fit into the free FANCI-FANCD2 interface. Free ID is rendered semi-transparent. b, Superposition of the open and closed conformation Core-bound ID complexes aligned on the middle-portion of the FANCI NTD. Top, overall view as in a, showing the rotation axes of the NTD-NTD interface (long gray line) and the FANCI N-terminal segment (short gray line) in the open-to-closed transition. Bottom, superposition viewed approximately along the NTD rotation axis, marked with circular arrow, also showing the FANCL’ RING. Open conformation ID is rendered semi-transparent. c, Top, superposition of the Core-bound closed-conformation ID and IDUb aligned on the mid-portions of their FANCI NTD domains. Also shown is the rotation/translation axis to bring the NTDs into register (top right line), and the rotation axes of the FANCI and FANCD2 N-terminal segments for the closed-conformation to IDUb transition (Extended Data Fig. 9d-f). Bottom, superposition viewed along the horizontal direction above, also showing the FANCL’ RING which would clash with the ubiquitin (yellow) attached to FANCD2 of IDUb, which is rendered semi-transparent.