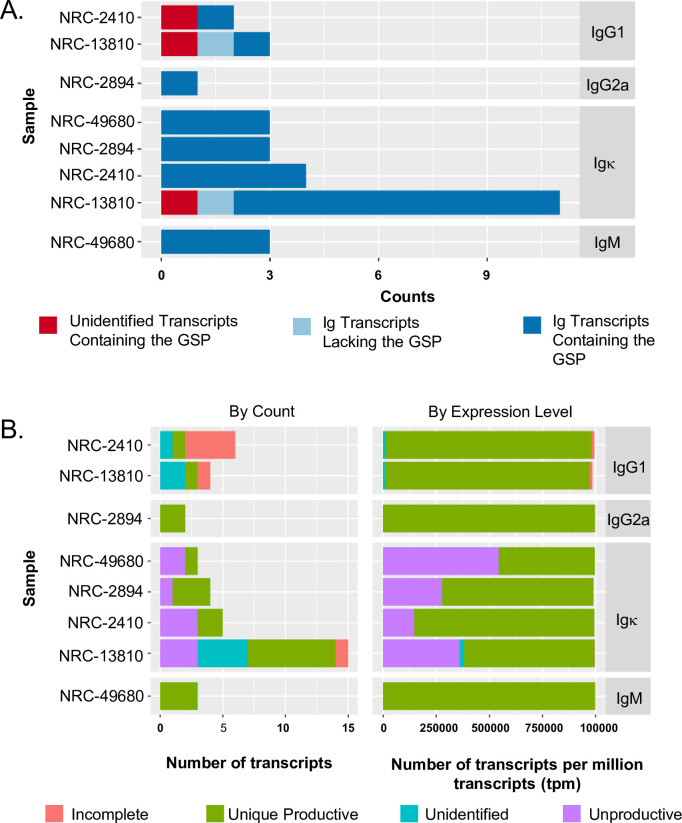
Fig 2. Hybridoma Ig sequencing using MiSeq Illumina NGS and de novo transcript assembly.
Profiles of Contig types among the 4 hybridomas. A. Summary of hybridoma sequence contig assemblies identified using isotype-specific ISPs. B. The Trinity de novo assembled Ig contigs were grouped and quantitated based on counts (Left) or expression levels (Right) into four functional categories: 1) Unique Productive—contigs without STOP codons in the CDRs and found in only one of the tested hybridomas, 2) Unproductive—contigs with one or more STOP codons in the CDRs, 3) Incomplete—only 1 or 2 CDRs identified, and 4) Unidentified—no CDRs identified.