Fig. 1. Architecture of DeepDEP.
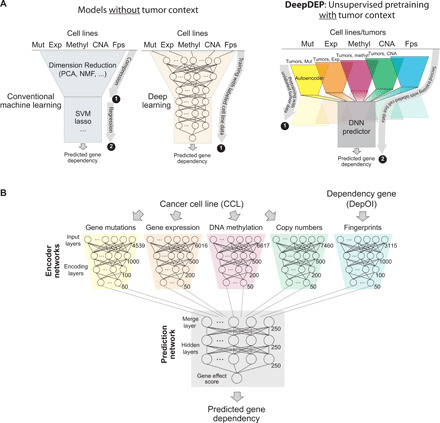
(A) Study designs of conventional methods and the proposed model with an unsupervised pretraining scheme. Conventional ML methods (left) were designed on the basis of merely labeled data of CCLs on related topics, such as prediction of drug sensitivity. An analog DL model can be implemented using a similar scheme (middle). The proposed DeepDEP model (right) has an unsupervised pretraining design that captures unlabeled tumor genomic representations and is further trained with gene dependency data of CCLs (labeled data). Mut, mutation; Exp, expression; methyl, DNA methylation; Fps, functional fingerprint of gene dependencies; DNN, deep neural network. (B) Architecture of DeepDEP. DeepDEP is designed to predict the gene effect score of a dependency of interest (DepOI) for a cancer sample (CCL or tumor). It is composed of encoders for each type of genomic data of a sample and fingerprint of a DepOI; the former was transferred from an unsupervised pretraining of autoencoders using tumors of TCGA. The complete model was trained and tested using CCL data of Broad DepMap.
