Fig. 2. Performance and validation of DeepDEP.
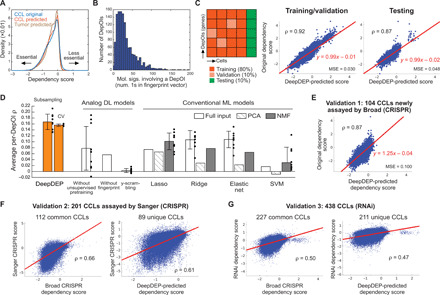
(A) Distributions of original dependency scores of CCLs and predicted scores of CCLs and tumors. (B) Nonzero elements in functional fingerprint vectors (membership of 3115 molecular signatures) of 1298 DepOIs. (C) Model performance of DeepDEP. (D) Performance comparisons with other DL and ML methods. Average per-DepOI ρ across 1298 DepOIs are summarized by means (bars) and SD (error bars) of 10 independent subsampling trainings (dots), or three rounds of 10-fold cross-validations (CV). The results were contrasted to identically structured DL models trained merely on CCLs without unsupervised pretraining on tumors, trained by individual DepOIs without the fingerprints to learn from other DepOIs, or with scrambled dependency scores (“y-scrambling”), and linear and nonlinear conventional ML methods with full or dimension-reduced inputs of genomic data and gene fingerprints. For each ML method, a model was trained using the same training/testing partition as used in the final reported DeepDEP model. Using the type of input data [full, principal component analysis (PCA), or nonnegative matrix factorization (NMF)] yielding the best performance for each ML method, we further trained and tested 10 models with the training/testing subsamples as used for DeepDEP (the first bar). Support vector machine (SVM) with nonlinear Gaussian and radial basis function (RBF) kernels did not converge and are not included here. A data point of SVM with NMF inputs at −0.07 and the SD of the model without the fingerprints at 0.23 are not shown since they are beyond the range of the vertical axis. Results of two subsets of highly variable DepOIs are shown in fig. S2. (E to G) Analysis of three independent validation datasets. Scores predicted by DeepDEP were compared to dependency scores in (E) newly assayed CCLs by Broad DepMap using a refined computational pipeline, (F) the Sanger DepMap using a different CRISPR library and computational algorithm, and (G) RNAi-based screens.
