Figure 6. Depletion of STAG2 enhances the migration and metastatic potential of Ewing sarcoma cells.
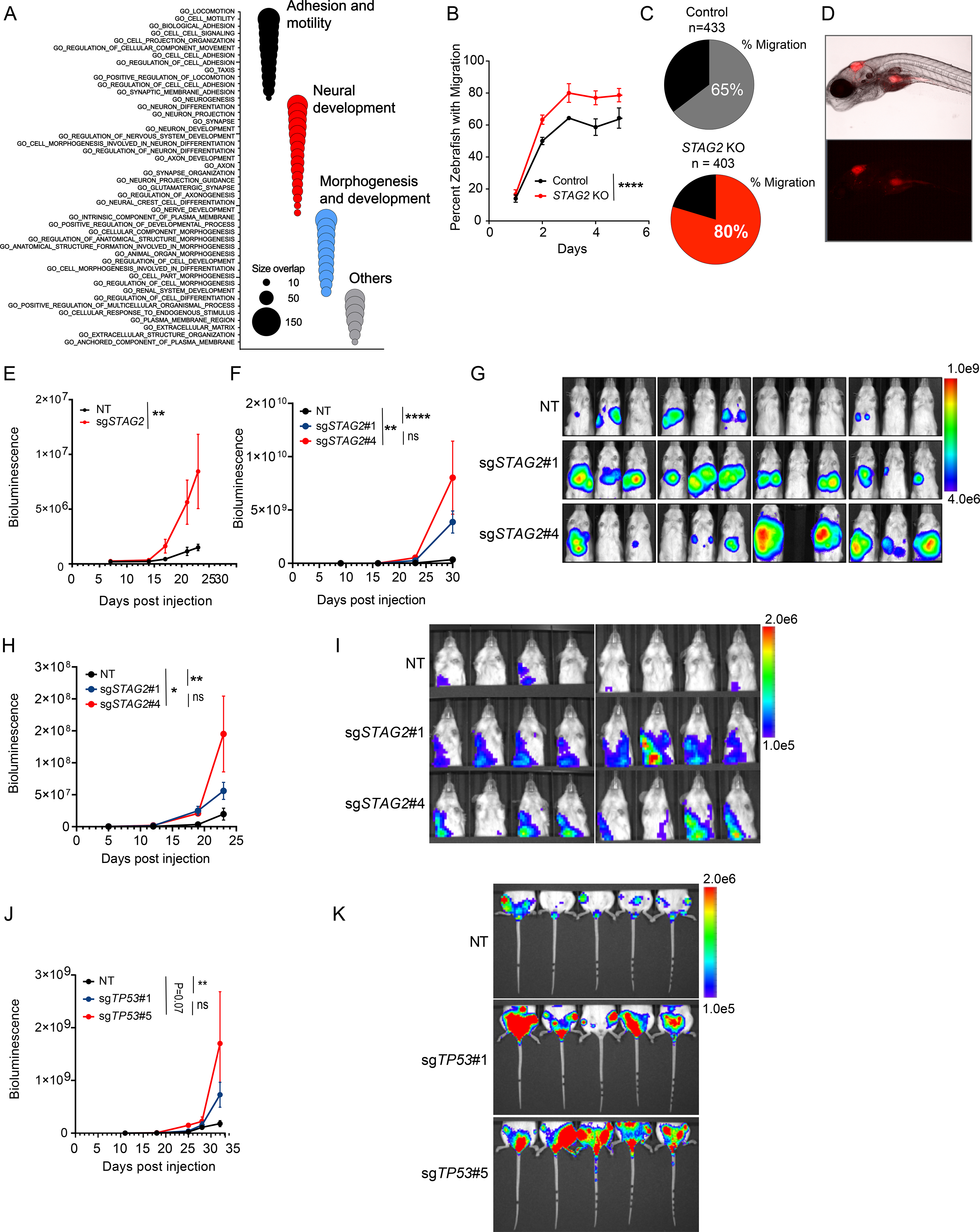
(A) Bubble plot summarizing top 50 significant enrichments (size overlap≥10, P≤0.05, FDR≤0.05) for the MSigDB v7.1 c5 collection. Enriched gene sets are clustered in representative functional categories. (B) Line graph represents mean ± sd of the percentage of zebrafish that displayed migration to at least one of the three regions of interest as a function of the days post injection. (2-way ANOVA, **** P < 0.0001). (C) Pie chart showing the percentage of zebrafish with migration of Ewing cells to at least one of the three sites examined three days post injection. (D) Representative brightfield (top) and fluorescence (541/565 nm bottom) images displaying migration of TC71 cells to the yolk sack. (E) Quantification of bioluminescence signal collected for lower extremities after blocking upper abdominal cavity. Line graph represents mean ± SEM, N = 8 per group. (2-way ANOVA, ** P<0.001). (F) Quantification of bioluminescence signal collected for upper thoracic cavity after blocking lower abdomen. Line graph represents mean ± SEM, N = 12 per group (2-way ANOVA, **** P < 0.0001,** P < 0.01, ns = not significant). (G) Bioluminescence images of mice described in Figure 6F. (H) Quantification of bioluminescence signal collected for upper thoracic cavity while blocking lower abdomen. Line graph represents mean ± SEM, N = 8 per group. (2-way ANOVA, **** P 0.0001,** P < 0.01, ns=not significant). (I) Bioluminescence images of mice described in Figure 6H. (J) Quantification of bioluminescence signal collected for lower extremities after blocking upper abdominal cavity. Line graph represents mean ± SEM, N = 5 per group. (2-way ANOVA, ** P < 0.01, ns = not significant). (K) Bioluminescence images of mice described in Figure 6J. See also Figure S6 and Table S7.
