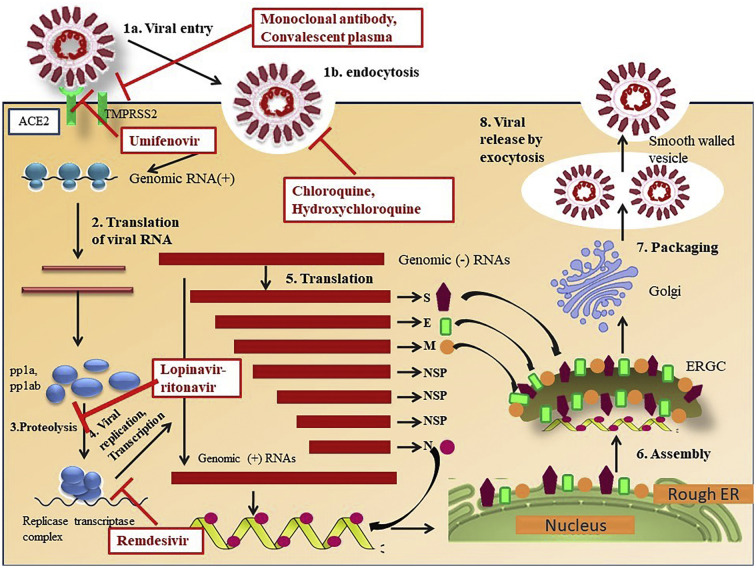
Fig. 2.
Molecular mechanism of relevant drugs and therapies for the infection of SARS-CoV-2. SARS-CoV-2 enters into the host cell via angiotensin converting enzyme 2(ACE2) receptor by endocytosis with the release of its RNA contents into the recipient cell to replicate. Monoclonal antibody or convalescent plasma therapy inhibits the ACE2 receptor or interferes with transmembrane serine protease 2 (TMPRSS2) function to affect the viral binding with the host cell. Chloroquine and hydroxychloroquine interfere with the entry of virus via endocytosis. Umifenovir targets spike (S) protein/ACE2 interaction and inhibits membrane fusion of the viral envelope. After the shedding of viral RNA, it translates into polypeptide (pp)1b, pp1ab leading to the formation of double membrane vesicles (DMVs) and establishment of the replication-transcription complex (RTC). This stage is followed by generation of intermediate negative strand RNA from which more numbers of positive strand RNA and mRNAs are generated. Lopinavir/ritonavir inhibits protease to prevent proteolysis, a mechanism that establishes RTC. Remdesivir targets the RNA dependent RNA polymerase and hamper the replication cycle of RNA viruses. Structural nucleocapsid (N) proteins are generated from the translation of nucleocapsid (N) mRNA, which in turn encapsulates the newly generated positive RNA strands. However, other structural proteins, envelope (E) proteins and membrane (M) proteins are formed via the translation in endoplasmic reticulum (ER) and gather in endoplasmic reticulum golgi intermediate complex (ERGIC) and cis-Golgi. In addition to the structural proteins some non-structural proteins (Nsps) are also generated. The assembly of the viral components begins when the accumulated proteins exit from the golgi apparatus and eventually fuse with the cell membrane, resulting in the release of new virus particles.