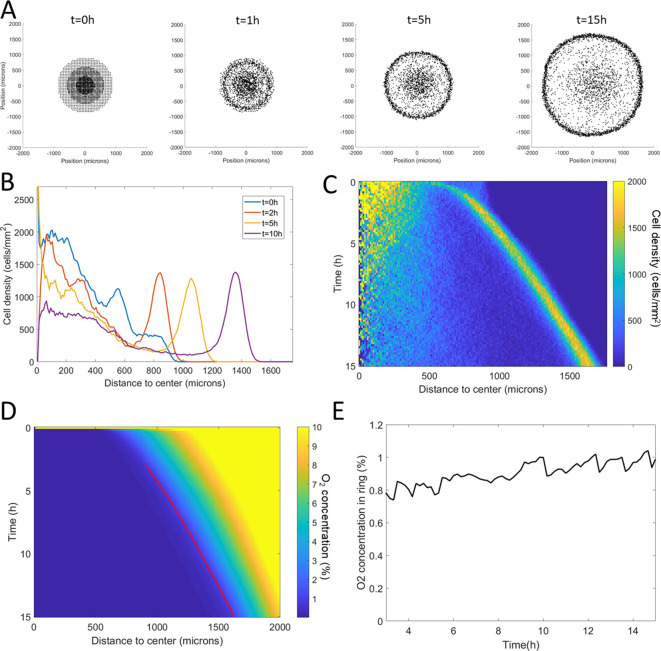
Figure 4. Minimal Potts model of ring formation and migration.
(A) Snapshots of a simulated colony of cells showing the formation of highly dense ring of cells. (B) Cell density profiles averaged over all angles for four different times. (C) Corresponding kymograph of cell density (colorbar in cells/mm2) as a function of time and distance to the center. Quantification in terms of microns and hours is described in the Materials and methods section. (D) Kymograph of O2 concentration (colorbar in %) with the position of the ring represented as a red line. The colormap is limited to the 0–10% range for readability but earlier time points show concentrations higher than the 10% limit. (E) O2 concentration at the ring position as a function of time showing that, here too, the ring follows a constant O2 concentration.