Figure 2.
Structural basis of MD65 binding to COVID-19 spike variants determined by comparative modeling to the CovA2-04 mAb
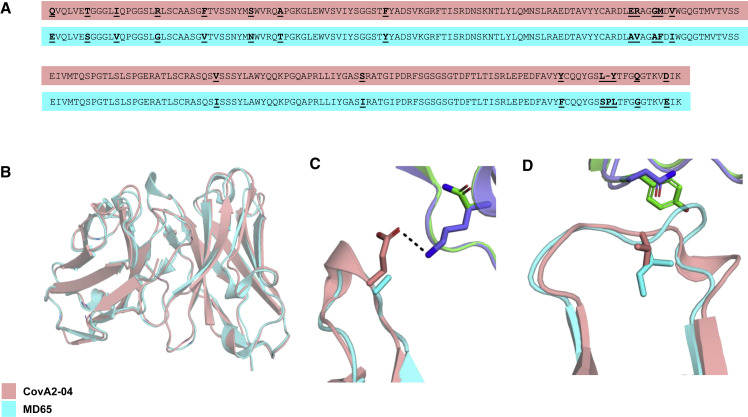
(A) Alignment of the primary amino acid sequences of the heavy (two upper sequences) and light (two lower sequences) chain variable domains of the two antibodies compared (CovA2-04 in pink and MD65 in cyan). Diverged residues are indicated by bold and underlined letters.
(B–D) MD65 model structure (cyan) aligned with CovA2-04 crystal structure (pink; WT [PDB: 7jmo] and B.1.351 spike [PDB: 7nxa], violet and green, respectively). (B) A view of the superimposed variable domain models of the two mAbs, indicating the close correspondence of the MD65 model structure and the experimentally determined structure of CovA2-04. (C) The Lys residue at position 417 of the WT spike protein forms a stabilizing hydrogen bond (dashed line) with the Glu residue at position 100 of CovA2-04. The K417N present in B.1.351 abrogates this stabilizing interaction leading to potential strain in binding to the negative surface on the complementarity-determining region (CDR) H3 loop of CovA2-04. The Ala at the analogous position in MD65 may relieve this strain. (D) The small-to-large N501Y substitution on the B.1.351 spike may physically overlap with the CDR L1 of CovA2-04. The V29I difference in MD65 (compared with CovA2-04) modifies the CDR L1 backbone conformation, expanding the space in this region for the bulkier Tyr residue of the spike.