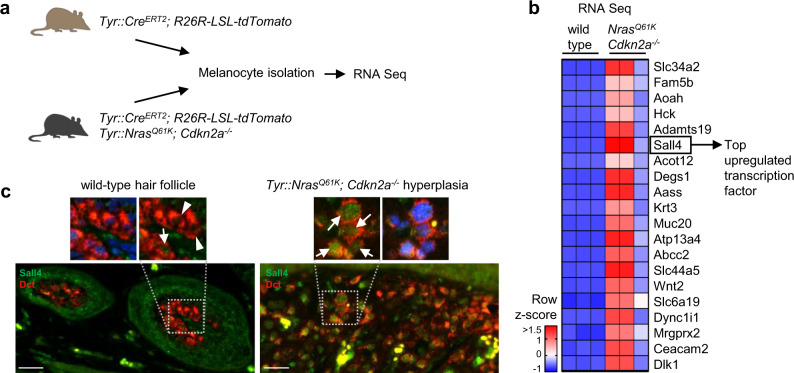
Fig. 1. Sall4 is upregulated in hyperplastic melanocytes.
a Experimental scheme of isolated melanocytes for RNA sequencing either from Tyr::CreERT2; R26R-LSL-tdTomato wild-type mice or Tyr::CreERT2; R26R-LSL-tdTomato; Tyr::NrasQ61K; Cdkn2a−/− mice with hyperplastic, melanoma-prone skin. At the age of 4 months, the mice were injected on 5 consecutive days with 80 µg g−1 body weight tamoxifen to induce tdTomato expression in melanocytes. The hair growth cycle in wild-type animals was synchronized by dorsal hair plucking. Melanocytes were isolated from mouse back skin of wild-type and melanoma model mice (3–6 mice per sample) and FACS sorted for cKit (staining) and tdTomato (endogenous). b Row z-score heatmap of normalized counts from RNA sequencing as illustrated in a. Visualized are the top 20 most upregulated genes comparing hyperplastic (Tyr::NrasQ61K; Cdkn2a−/−) to wild-type melanocytes. Genes are ordered top to bottom with genes with highest LogFC values on top. Cutoffs were set at p values < 0.05 and FDR < 0.05. Sall4 (LogFC value = 9.59) represents the only and hence highest upregulated transcription factor within those top 20 upregulated genes. The complete list of all differentially expressed genes can be found under Supplementary Data 1. c Immunohistochemical staining of wild-type melanocytes in murine hair follicles (left panel) and melanoma-prone melanocytes in hyperplastic Tyr::NrasQ61K; Cdkn2a−/− mouse skin (right panel). Arrowheads point towards wild-type melanocytes with undetectable Sall4 expression; arrows point towards hyperplastic and rare wild-type melanocytes with detectable Sall4 protein. Experiment has been repeated independently three times with similar results. Scale bars 25 µm.