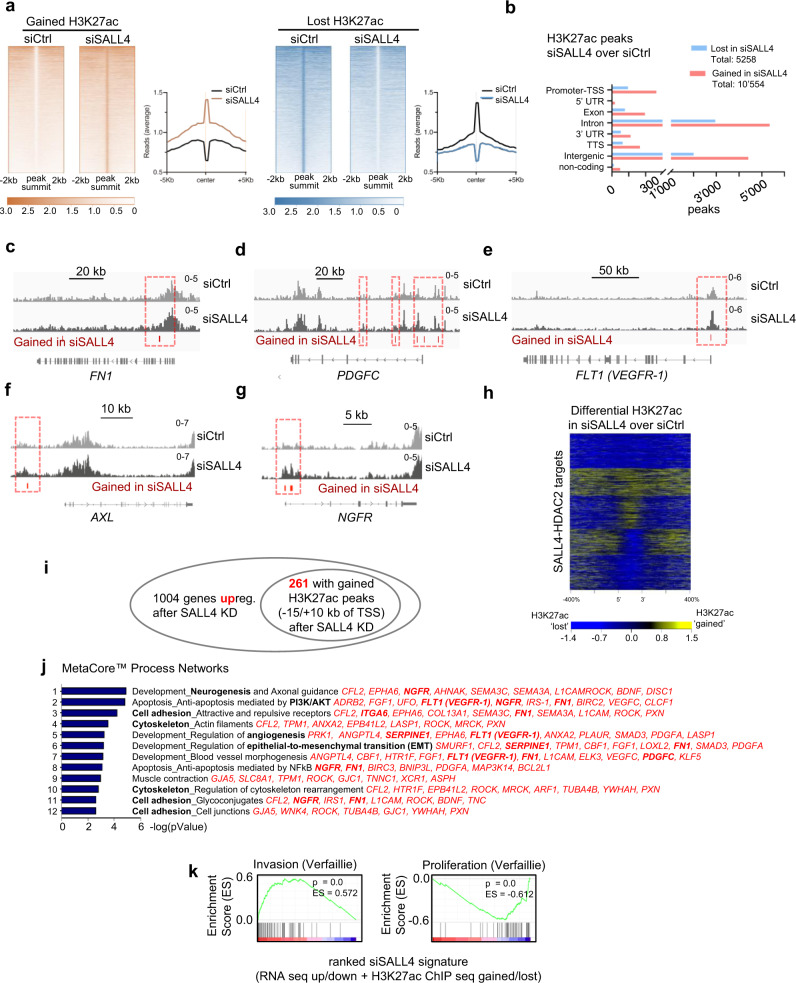
Fig. 7. SALL4 can regulate invasiveness genes through an epigenetic mechanism.
a Read density heatmap of gained and lost H3K27ac ChIP sequencing peaks (±2 kb) upon SALL4 knockdown (left panels) and average read distribution of gained and lost H3K27ac peaks within ±5 kb from peak center (right panels). b Distribution of the gained and lost H3K27ac peaks in different genetic regions. TTS: transcription termination site, TSS: transcription start site, UTR: untranslated region. c–g Representative tracks of invasiveness genes with significantly gained (red bars; highlighted by red, dashed lines) H3K27ac peaks in siSALL4 over siCtrl (no significantly lost H3K27ac peaks were detected for the visualized gene tracks). h Read density heatmap of differential H3K27ac ChIP-seq peaks (within 10 kb) in siSALL4 over siCtrl at direct target genes of SALL4-HDAC2 (CUT&RUN peaks with at least 3 of 4 SALL4/HDAC2 antibodies). i Significantly upregulated genes after SALL4 knockdown (Fig. 4a, b) were overlaid with those genes that have significantly gained H3K27ac marks −15/+10 kb of TSS (a, ocher panel). This resulted in 261 genes with activating chromatin marks that were at the same time upregulated after SALL4 knockdown. j MetaCore™ Process Network enrichment of the 261 genes from i. The top 12 most significant processes are indicated with the specific genes of each process listed in red. k Gene Set Enrichment Analysis (GSEA) of the combined 261 upregulated (i) and 137 downregulated genes (Supplementary Fig. 17a) with differential acetylation after SALL4 knockdown (ranked according to log2 expression ratio of RNA seq results) with the previously published melanoma programs of Verfaillie and colleagues (2015). Source data for b are provided as a Source Data file.