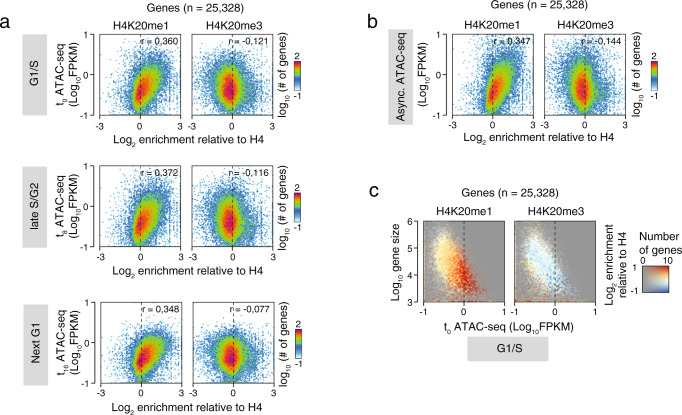
Fig. 1. Chromatin accessibility at genes bodies correlates with histone H4 lysine 20 mono-methylation throughout the cell cycle.
a 2D-histograms showing the relationship between enrichment of selected H4K20 methylation states (X-axis) and chromatin accessibility probed using ATAC-seq (Y-axis) at human genes. X-axis shows log2 normalized H4K20me1 (left side) or H4K20me3 (right side) ChIP-seq levels relative to the levels of H4 control ChIP-seq, while Y-axis shows log10-transformed average FPKM-normalized ATAC-seq signal from synchronized U2OS cells at different cell cycle phases, i.e., G1/S (0 h—top, three replicates), S-G2 (8 h—middle, three replicates) and next G1 phase (16 h—bottom, four replicates). The color scales reflect the number of genes having a certain combination of H4K20me enrichment and ATAC-seq. r-values show the Pearson’s correlation coefficients for the data in each plot. b 2D-histograms showing the relationship between enrichment of selected H4K20 methylation states (X-axis) and chromatin accessibility probed using ATAC-seq (Y-axis) at human genes as in a. Y-axis shows log10-transformed average FPKM-normalized ATAC-seq signal from four replicates of U2OS cells grown asynchronously. c Colored 2D-histograms showing the three-way relationship between enrichment of selected H4K20 methylation states (color), ATAC-seq signal (X-axis), and gene sizes (Y-axis) at human genes. Coloring shows log2 normalized H4K20me1 (left side) or H4K20me3 (right side) ChIP-seq levels relative to the levels of H4 control ChIP-seq with the number of genes with a certain combination of ATAC-seq signal and size controlling opacity as indicated in the right side color scale. X-axis show log10-transformed average FPKM-normalized ATAC-seq signal from three replicates of synchronized U2OS cells 0 h after release.