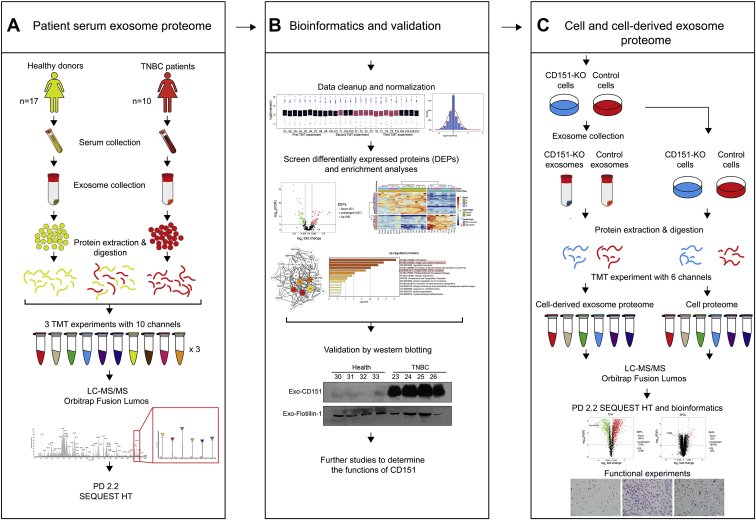
Fig. 1.
Workflow for characterization of TNBC-related changes in serum and cell line–derived exosome proteome.A, five hundred microliter of serum was collected from ten patients with TNBC and 17 healthy donors, and exosomes were purified by ultracentrifugation. Exosomes were lysed, and 100 μg of exosomal protein was digested. The peptides in each individual sample and the pooled samples were randomly distributed and labeled across three 10-plex TMT experiments. After labeling, samples were mixed within each TMT experiment and separated into seven fractions. Tandem MS data were obtained using an Orbitrap Fusion Lumos instrument, and proteins were identified using Proteome Discoverer 2.2. B, the serum exosome proteome data were normalized and analyzed. DEPs were determined by t test and further characterized through gene enrichment and network analysis. Representative protein CD151 was validated by Western blotting. C, CD151 was knocked out in MDA-MB-231 cells by using CRISPR–Cas9. Quantitative proteomics approach was performed to reveal the proteomes of CD151-deleted exosomes and cells. DEP, differentially expressed protein; TMT, tandem mass tag; TNBC, triple-negative breast cancer.