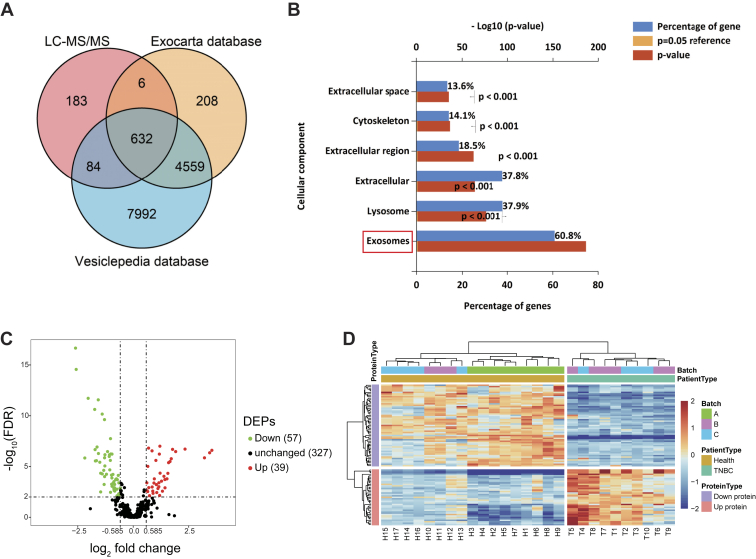
Fig. 4.
Global profiling of proteins encapsulated in TNBC-derived exosomes using MS.A, Venn diagrams of proteins identified in TNBC-derived exosomes by MS. The total number of proteins identified was compared with published results for exosomal and extracellular vesicle proteins from the Exocarta and Vesiclepedia databases. B, intracellular locations of all identified proteins assigned by the FunRich 3.1.3 tool. C, volcano plot of quantified proteins constructed from log2 fold change (x-axis) and −log10 q value (y-axis) data. The threshold for determining DEPs is indicated by the dashed lines (adjusted p value < 0.01, fold change > 1.5). Red dots indicate significantly upregulated proteins, and green dots indicate significantly downregulated proteins in TNBC serum exosomes. D, hierarchical clustering analysis of a heat map of the 96 DEPs was performed based on Euclidean distance by the R package “pheatmap,” revealing that the molecular profile of each group is unique, with the individuals in each group being closest together. The red color indicates increased protein abundance, and the blue color indicates decreased protein abundance. DEP, differentially expressed protein; TNBC, triple-negative breast cancer.