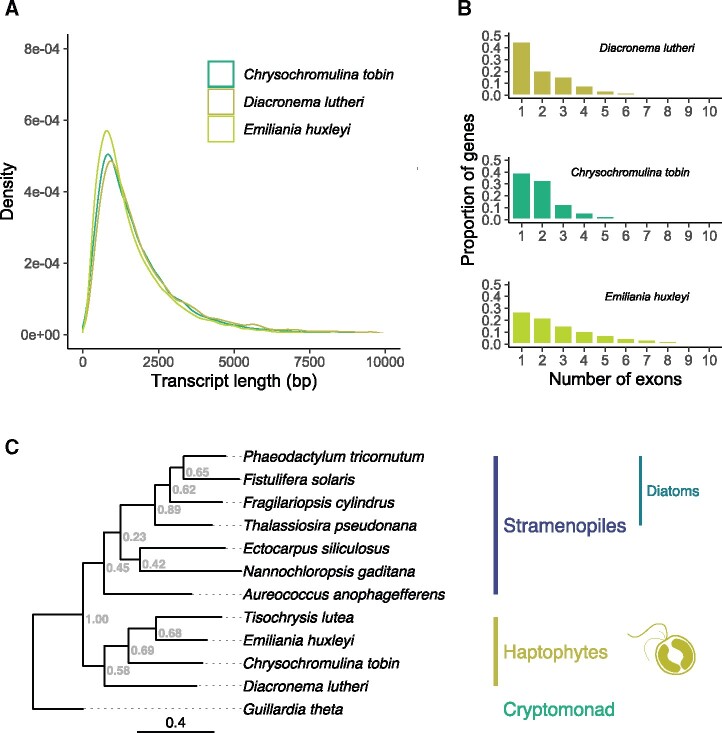
Fig. 1.
(A) Gene transcript length distributions for Diacronema lutheri annotated in this study compared with two other haptophytes, Emiliania huxleyi and Chrysochromulina tobin. (B) The number of exons per gene for the same three genomes, expressed as a proportion of the total number of annotated genes. (C) The species tree of 12 protists bearing red alga derived plastids, including four sequenced haptophytes, seven stramenopiles, and one cryptomonad. The tree is derived from 1,152 orthogroups with at least one gene copy from each species. Branch lengths represent substitutions per site and support values are derived from the STAG algorithm implemented in OrthoFinder2.