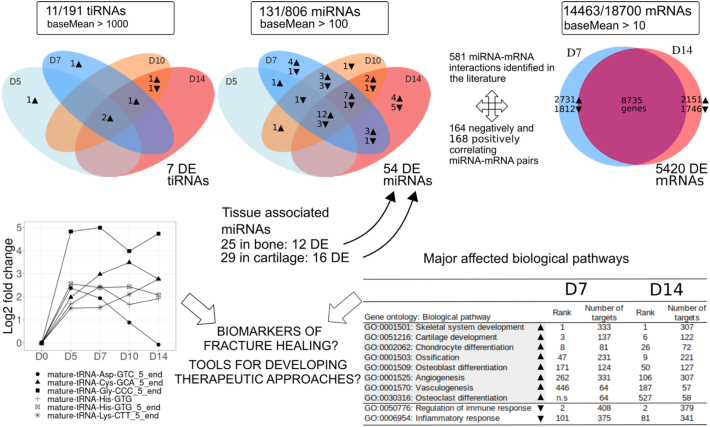
Fig. 7.
Schematic summary of the genome wide expression analysis of tiRNAs, miRNAs and mRNAs during early fracture healing in mice. Cutoff for the presented values, average normalized read count (baseMean) for tiRNAs was >1000, for miRNAs > 100 and for mRNAs > 10. Values in boxes indicate the number of genes with expression level above the baseMean cutoff, and total number of identified genes. Number of bone and cartilage associated miRNAs are presented. Differential expression (DE) (with an absolute value of log2FC > 2 against 2 M diaphyseal bone (D0) baseline and Padjusted-value < 0.05) is illustrated at D5, D7, D10 and D14 for tiRNAs and miRNAs and at D7 and D14 for mRNAs. The gene ontology analysis was based on mRNA expression data at D7 and D14. ▼: downregulation; ▲: upregulation. Rank associated with GO terms have P-value < 0.05. Grey color highlights the upregulated biological GO pathways.