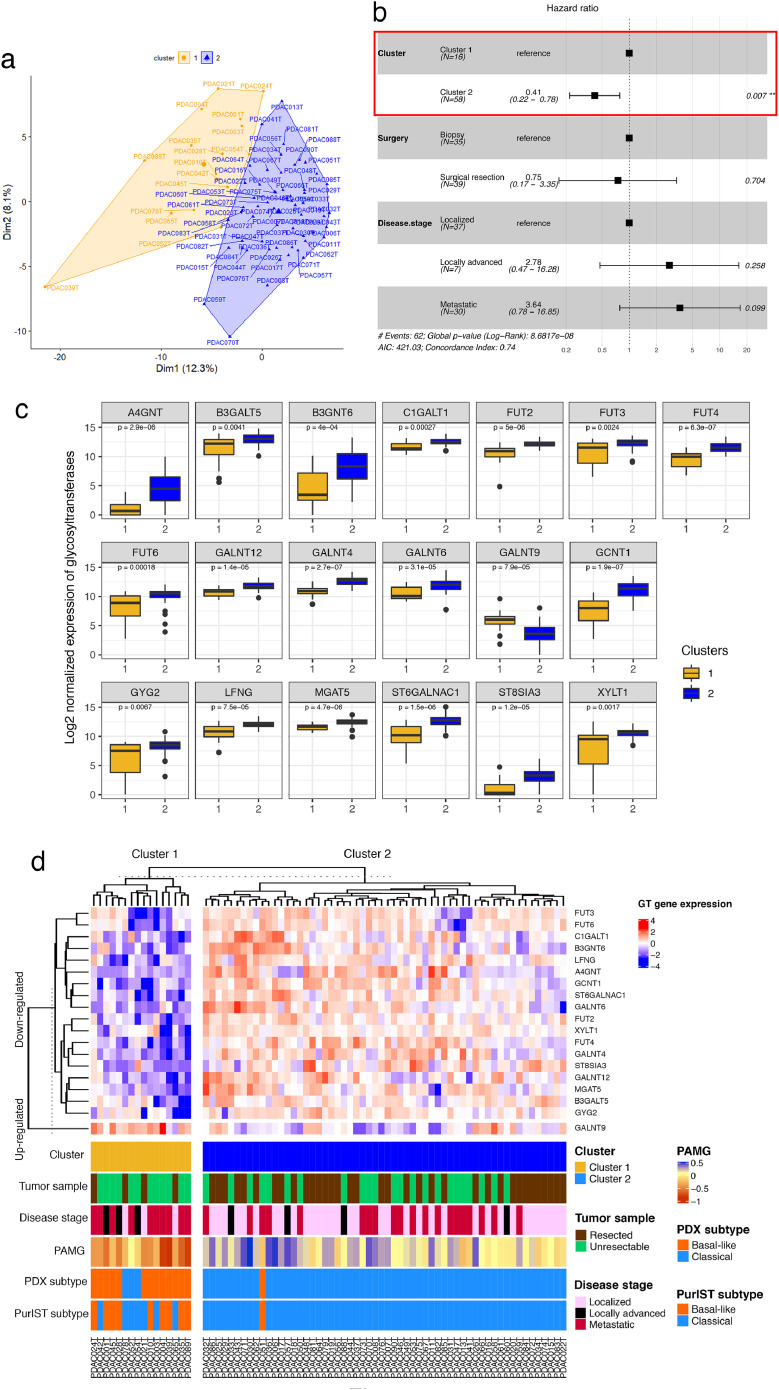
Fig. 1.
Prognostic stratification of PDAC through their GT gene expression profile (a, b) and clinical features of patients with their PDAC molecular profiles (c, d). (a) Biplot of the HCPC analysis result based on RNA-seq data of 74 PDAC and 169 GT genes. (b) Forest plot of the multivariate survival analysis including PDAC clustering, surgical resection and the disease stage. (c) Boxplot showing Log2 normalized expression of 19 GT genes stratified by clusters 1 and 2. The lower and upper hinges correspond to the first and third quartiles (the 25th and 75th percentiles). Kruskal-Wallis p-value was used for the statistical significance between clusters. (d) Heatmap of the two clusters based on transcription signature of 19 GT genes. Clinical characteristics and molecular classifications of each patient tumor were shown in the corresponding annotation. Color bars indicate the tumor sample type, disease stage, PAMG and basal-like/classical subtyping based on previously established consensus [30] and PurIST classifier.