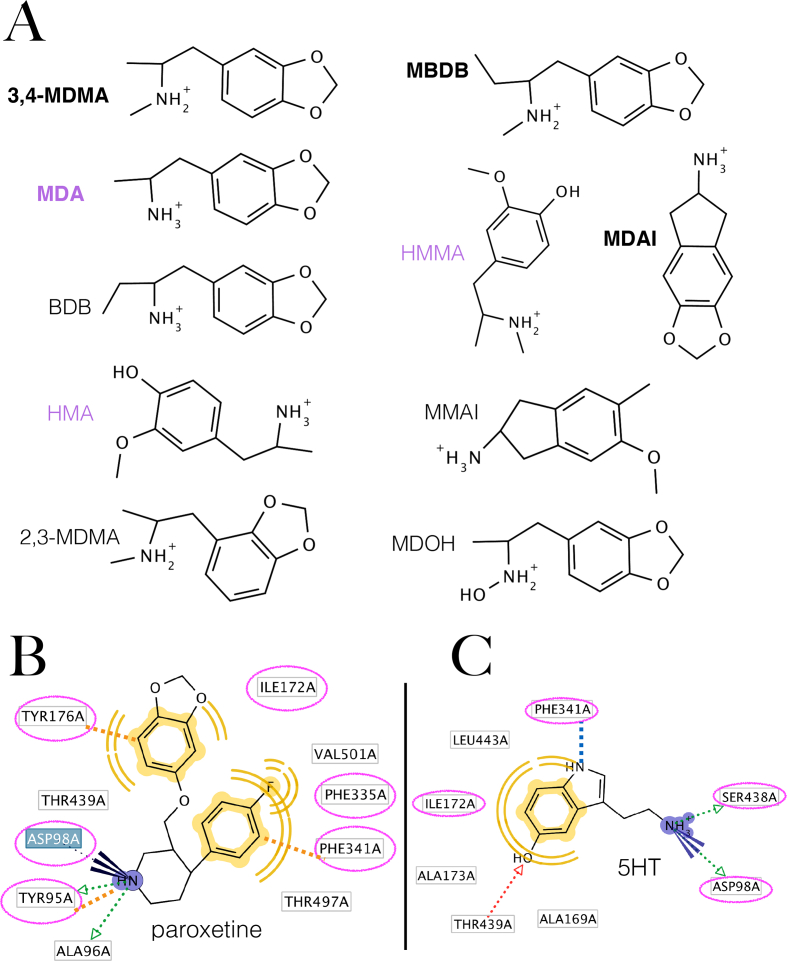
Figure 1.
Molecules chosen to generate the pharmacophore and 2D representation of the binding mode of paroxetine and serotonin (A) Selected SERT active, MDMA congeneric compounds to design the structure-based pharmacophore. Molecules in bold are the first described entactogens (Nichols, 1986) and compounds in purple are primary metabolites of MDMA. (B) Pharmacophore of paroxetine to the hSERT calculated from its intact observed complex PDB: 6VRH. Computed residues that also bind MDMA (vide infraFigure 3B) are in circles in magenta. (C) Pharmacophore of serotonin on the hSERT generated in the present work. Discontinuous blue line indicates a cation-π interaction, dotted green arrows represent Hbond (donors), blue rays represent an ionic interaction, yellow moieties and waves represent hydrophobic interactions and discontinuous orange lines indicate a cation-π with Y95 and two edge-to-face π-π interactions with Y176 and F341.