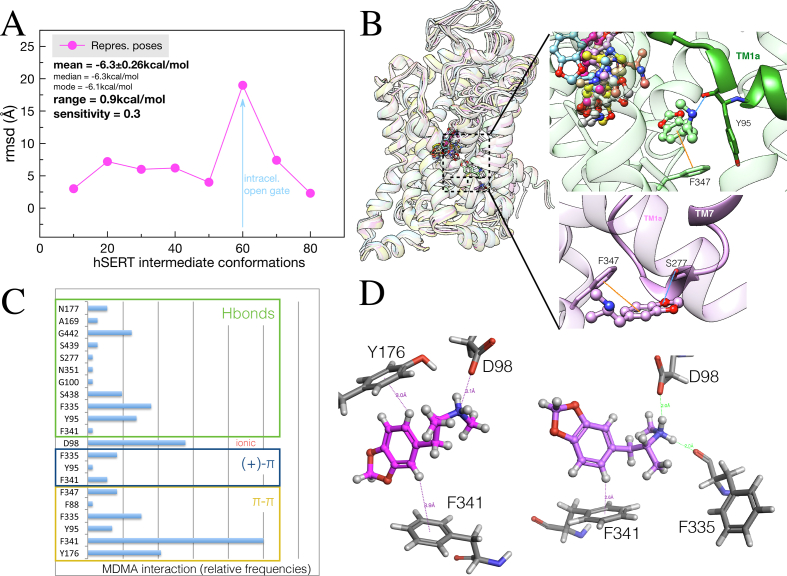
Figure 5.
Ensemble binding space analysis of MDMA on the hSERT. (A) Parameterized ‘Ensemble binding space docking’ of MDMA on interpolated trajectories between two hSERT outward-open, one occluded and an inward-open structure (PDBs: 5I71, 6VRH, 6DZV and 6DZZ). Data points (in magenta) represent the rmsd values between the most populated representative docking solutions at each intermediate model. The blue arrow represents the migration of MDMA towards the open gate at the cytoplasmic side. (B) Representative MDMA/hSERT binding complexes, the inserts show the interactions of the most populated binding solutions closest to the inward gate. π-π stackings are in orange and in Hbonds in blue. (C) Relative frequencies of the residues interacting electronically with MDMA along the interpolated trajectories, i.e. calculated from representative binding poses from dockings simulations carried out in 1 every 10 intermediate conformations. Every docking solution (N = 80) was clustered into 8 representative biding poses. (D) Two representative binding poses illustrating the most frequent interactions.