Significance
The topology of membrane proteins is defined by the successive integration of α-helical transmembrane domains at the Sec61 translocon. For each polypeptide segment of ∼20 residues entering the translocon, their combined hydrophobicities were previously shown to define membrane integration. Here, we discovered that different sequences preceding a potential transmembrane domain substantially affect the hydrophobicity threshold. Sequences that are rapidly folding, intrinsically disordered, very short, or strongly binding chaperones allow efficient integration at low hydrophobicity. Folding deficient mutant domains and artificial sequences not binding chaperones interfered with membrane integration likely by remaining partially unfolded and exposing hydrophobic surfaces that compete with the translocon for the emerging transmembrane segment, reducing integration efficiency. Rapid folding or strong chaperone binding thus promote efficient integration.
Keywords: membrane proteins, molecular chaperones, protein folding, Sec61 translocon, topogenesis
Abstract
The topology of most membrane proteins is defined by the successive integration of α-helical transmembrane domains at the Sec61 translocon. The translocon provides a pore for the transfer of polypeptide segments across the membrane while giving them lateral access to the lipid. For each polypeptide segment of ∼20 residues, the combined hydrophobicities of its constituent amino acids were previously shown to define the extent of membrane integration. Here, we discovered that different sequences preceding a potential transmembrane domain substantially affect its hydrophobicity requirement for integration. Rapidly folding domains, sequences that are intrinsically disordered or very short or capable of binding chaperones with high affinity, allow for efficient transmembrane integration with low-hydrophobicity thresholds for both orientations in the membrane. In contrast, long protein fragments, folding-deficient mutant domains, and artificial sequences not binding chaperones interfered with membrane integration, requiring higher hydrophobicity. We propose that the latter sequences, as they compact on their hydrophobic residues, partially folded but unable to reach a native state, expose hydrophobic surfaces that compete with the translocon for the emerging transmembrane segment, reducing integration efficiency. The results suggest that rapid folding or strong chaperone binding is required for efficient transmembrane integration.
Transmembrane α-helices are the basic structural principle of most membrane proteins. As a result, the topology of multispanning membrane proteins is defined by a succession of helices of alternating orientation, separated by loops exposed to the cytoplasm and to the exoplasmic space [i.e., the lumen of the endoplasmic reticulum (ER) in eukaryotes or the exterior of the plasma membrane in prokaryotes (1)]. Upon integration, the transmembrane segments assemble to a helix bundle, while the loops fold in either the cytosol or the exoplasmic space, where chaperones may assist to prevent misfolding and aggregation.
The integration of transmembrane domains and translocation of exoplasmic loops are mediated by the conserved Sec61/SecY translocon composed of Sec61-αβγ in the ER of eukaryotes and SecYEG in prokaryotes (2). The main subunit Sec61-α/SecY is a 10-transmembrane helix bundle that can open a pore across the membrane for the translocation of hydrophilic polypeptide chains and a lateral gate for the exit of hydrophobic segments into the lipid environment. In its idle state, the translocon is closed and stabilized by a short helix that forms an exoplasmic plug and by a central constriction.
Proteins are targeted to the membrane by a hydrophobic signal sequence that, as it emerges from the ribosome, binds to signal recognition particle (SRP). The signal is either an amino-terminal cleavable signal peptide or the first transmembrane domain of the protein acting as an uncleaved signal anchor. SRP targets the translating ribosome to the SRP receptor and to the translocon. The ribosome binds to cytoplasmic loops of the C-terminal, half of the translocon, leaving a gap open toward the cytosol (3). As the protein is synthesized, its transmembrane segments are inserted successively into the lipid bilayer.
Three distinct membrane integration processes can be distinguished (schematically shown in Fig. 1A). First, the signal (anchor) activates the translocon by intercalating between the gate helices and exiting toward the lipid phase (2). In the process, the hydrophilic flanking sequence is inserted into the pore for transfer into the lumen, and the plug is pushed away. This has been elucidated by a number of structures of translocons engaged with signal sequences (3–5). Which end of a signal anchor will be translocated is mainly determined by the flanking charges [the “positive–inside rule” states that the more positive end will be cytosolic (6–8)] and the folding state of the N-terminal domain hindering translocation (9). As the nascent chain is moving through the translocon pore, a hydrophobic transmembrane segment stops further transfer by leaving via the lateral gate into the lipid membrane as a stop-transfer sequence. The following polypeptide is then synthesized through the gap between ribosome and translocon into the cytosol until a further transmembrane segment again engages with the translocon, exits into the membrane, and inserts the downstream sequence into the pore. This reintegration process is similar to the initial signal integration. Furthermore, alternating stop-transfer and reintegration events may account for any number of transmembrane domains.
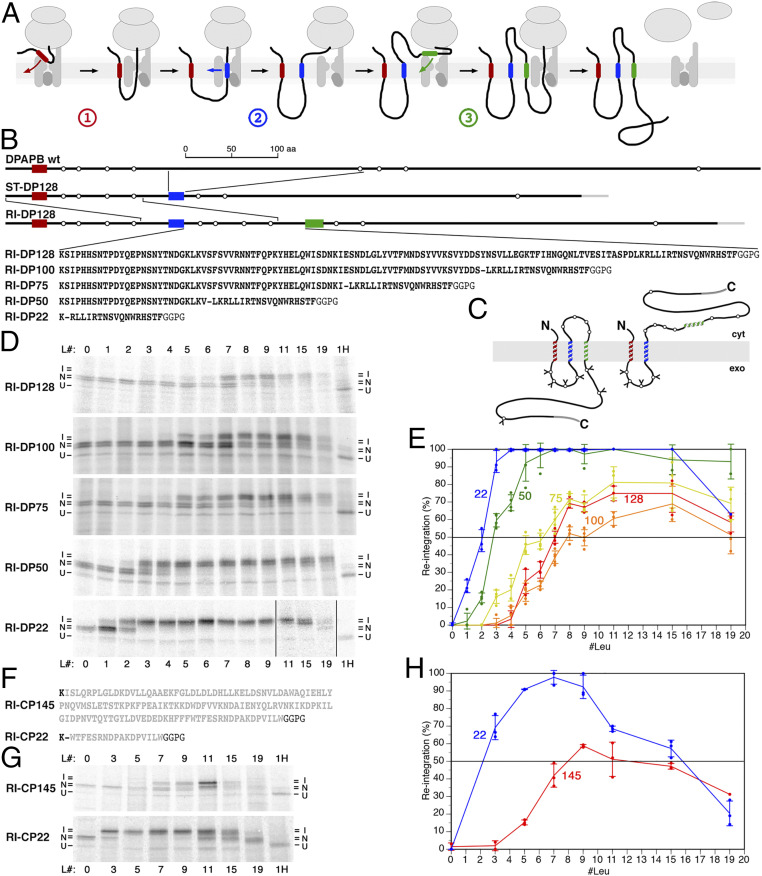
Fig. 1.
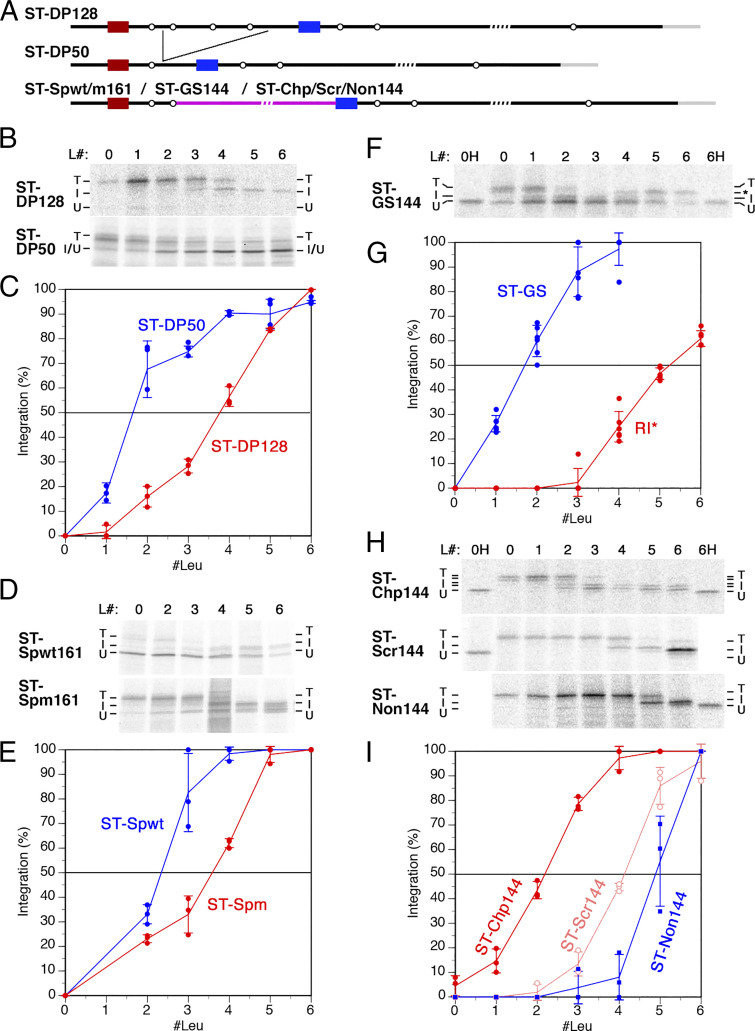
Reintegration efficiency is affected by the cytoplasmic upstream sequence. (A) Schematic representation of the three distinct integration processes in the biogenesis of membrane proteins: signal (anchor) integration (red transmembrane segment) (1), stop-transfer integration (blue) (2), and reintegration (green) (3). Transmembrane domains do not necessarily fully enter the translocon, before exiting into the membrane, but may associate with lipids early on and glide along the gate, as proposed by Cymer et al. (43). (B) The construct RI-DP128 was derived from wt DPAPB via ST-DP128 (DPAPB-H in ref. 12), as shown schematically with the transmembrane domains in red, blue, and green as in A. Rings indicate potential N-glycosylation sites. A C-terminal triple–HA-tag is shown in gray. To analyze the hydrophobicity threshold of membrane integration, the H segment sequence GGPGAAAAAAAAAAAAAAAAAAAGPGG, with 0 to 19 alanines replaced by leucines, were inserted as a potential reintegration sequence (green) in RI-DP. Below, the DPAPB sequences of the cytosolic loop between the stop-transfer and the reintegration domains of RI-DP128 and the deletion constructs RI-DP100–22 are listed, with a dash indicating the deletion site. At least 20 residues preceding the reintegration H segment were kept constant in all constructs. (C) Schematic representation of the two topologies when the H segments do or do not initiate reintegration and C-terminal translocation. cyt, cytosol; exo, exoplasm = ER lumen. Glycosylations are indicated by Y. (D) The various constructs, each with different reintegration H segments composed of 19 alanines, with 0 to 19 of them replaced by leucine residues (L#), were expressed in yeast cells, labeled with [35S]methionine for 5 min, immunoprecipitated, and analyzed by SDS–gel electrophoresis and autoradiography. Based on the glycosylation pattern, integration (I) or nonintegration (N) of the H segment, as well as uninserted unglycosylated products (U) were distinguished. To identify the position of the unglycosylated polypeptide, a sample was analyzed after deglycosylation by endoglycosidase H (H) on the right. (E) The fraction of products with reintegrated H segments as a percentage of the total membrane-integrated proteins was plotted versus the number of leucine residues in the H segment (mean, SD, and the individual values of at least three independent experiments). The curves are labeled with the length of the constructs’ cytosolic loops between stop transfer and H segment. (F) The cytoplasmic sequence between stop transfer and H segment is shown for RI-CP145 (residues 21 to 160 of pre-CPY, in gray) and RI-CP22 (residues 144 to 160). (G) The CPY constructs were expressed and analyzed, as in D. (H) Reintegration of the H segment was quantified, as in E (mean, SD, and the individual values of at least three independent experiments).
The main characteristic of transmembrane segments is their overall hydrophobicity on a length of ∼20 residues that are required for an α-helix to span the apolar core of a bilayer. This was confirmed by systematic quantitative analysis of a large number of so-called H segments, mildly hydrophobic sequences of 19 residues, for their efficiency as stop-transfer sequences (10). From these experiments, a “biological hydrophobicity scale” for all 20 amino acids was derived. The dependence of integration efficiency on hydrophobicity is consistent with a purely thermodynamic equilibration process between the pore interior and the lipid environment for each peptide segment entering the translocon. This concept allowed for the estimation of apparent free energy contributions ∆Gaaapp of each amino acid to membrane integration. The contribution of particular amino acids also depended on their position in a manner reflecting the structure of the membrane (11): Polarizable residues, such as tryptophan or tyrosine, energetically favor the polarity interface between apolar fatty acyl and polar lipid head group regions, while polar and charged residues are energetically most unfavorable in the center of the bilayer, thus reducing integration efficiency.
The equilibration model is further supported by mutagenesis of the translocon. The six residues forming the central constriction of the translocation pore are predominantly hydrophobic. Their mutation to more hydrophilic amino acids, to increase polarity and hydration inside the pore, reduced the hydrophobicity threshold for membrane integration considerably (12, 13), as expected for a partitioning process.
From these findings emerges the concept that each sequence of ∼20 residues, based on its amino acids and their position within the segment, autonomously defines its propensity to integrate into the membrane. The data on the energetics of membrane integration was mostly collected for stop-transfer integration at the mammalian ER. Yet similar hydrophobicity dependence and hydrophobicity scales for integration were determined in other systems, such as for stop transfer in yeast (12, 14) or bacteria (15, 16), for reintegration in yeast and mammalian ER (17) and even for TIM23-mediated integration into the inner mitochondrial membrane (18). The biological hydrophobicity scale thus is considered to be universal and generally applicable to transmembrane segments, irrespective of their orientation, the organism, or the compartment. Accordingly, it is also used for topology prediction from primary sequences (19).
However, the different systems that were analyzed also showed significant quantitative shifts in the hydrophobicity threshold for integration. A simple hydrophobicity series, as initially introduced by Hessa et al. (10), is produced with H segments consisting of 19 alanines, of which 0 to 19 are replaced by leucine. The threshold for 50% stop-transfer integration was determined to be 3.1 leucines using mammalian in vitro translation with dog pancreas microsomes and 3.8 leucines in baby hamster kidney cells (10). Further studies (summarized in ref. 1) measured thresholds between one and seven leucines [for example, 4.4 and 3.5 leucines in yeast (12, 14), 1.1 and 2.0 in Escherichia coli (15, 16), and 7.3 in HeLa cells (20)]. Using the ∆Gapp values from Hessa et al. (11), this covers a range of ∼3 kcal/mol. Different lipid compositions and translocon sequences, but also the reporter constructs, may contribute. As to the latter, it has indeed been observed that the sequence immediately following an H segment can influence integration efficiency, most likely by conformationally hindering or facilitating stop-transfer release into the bilayer (21). This already challenged to some extent the autonomy by which each sequence determines its integration propensity.
There is only one study that analyzed the hydrophobicity threshold of reintegration sequences (17). Surprisingly, it was found to be considerably lower than for stop-transfer integration with 0.9 versus 3.1 leucines in a mammalian in vitro translation/membrane insertion system and with 2.2 versus 4.4 leucines in yeast cells. In the present study, we set out to test the reintegration efficiency in Saccharomyces cerevisiae using the same H segments in a different reporter construct. Our results revealed a surprising dependence of reintegration on the length and/or characteristics of the sequence preceding the reintegration domain. A similar dependence was also found for stop-transfer integration. The data indicate that the folding properties of the cytoplasmic and luminal loop sequences in membrane proteins determine the integration efficiency of subsequent transmembrane domains.
Results
Reintegration Efficiency Depends on the Upstream Cytosolic Sequence.
To analyze the hydrophobicity threshold for reintegration, we employed a reporter derived from a preexisting stop-transfer construct (here called ST-DP128; Fig. 1B); the H segment was inserted into the yeast type II membrane protein dipeptidylaminopeptidase B (DPAPB; Dap2p), replacing residues 170 to 378. With this construct, the hydrophobicity threshold for 50% stop transfer had been determined to be 3.9 leucines (21). For the reintegration construct RI-DP128, the N-terminal 146 residues, including the signal anchor, were duplicated, such that the second copy of the signal anchor acts as a stop-transfer sequence, the following segment of 128 residues loops out into the cytosol, and the subsequent H segment may reintegrate and bring about the translocation of the C-terminal portion of DPAPB (Fig. 1C).
Constructs with different H segments were expressed in yeast cells and metabolically labeled with [35S]methionine for 5 min to detect the immediate translation and membrane integration products. Proteins were immunoprecipitated and analyzed by SDS–gel electrophoresis and autoradiography (Fig. 1D, Top and E, red line). The membrane topology of the products was derived from the glycosylation pattern relative to the deglycosylated protein after endoglycosidase H digestion. Constructs with zero to four leucines in their H segments produced a double band of three and fourfold glycosylated proteins corresponding to a twice membrane–spanning topology, with only a translocated loop between signal-anchor and stop-transfer sequences. With increasing hydrophobicity of the H segment, an additional double band appeared, consistent with six and sevenfold glycosylated products that also have their C terminus translocated, reflecting the H segment’s increasing reintegration activity. For 50% reintegration, seven leucines were required, much more than for stop transfer (3.9) (21) and than had been reported for reintegration before (2.2) (17). Unexpectedly, reintegration maximally reached 80% with 11 and 15 leucines and even declined to 60% with a maximally hydrophobic 19-leucine H segment. These results conflict with the notion that H segment integration is independent of sequence context.
A striking difference between our reporter construct and that of Lundin et al. (17) is that the length of the cytosolic loop preceding the reintegration H segment was much shorter with only 43 residues. To test the role of the cytoplasmic loop, we gradually shortened it in our reporter construct from 128 to 22 residues (Fig. 1 D and E). While reducing the loop size to 100 residues even lowered reintegration efficiency, it strongly increased from 75 to 22 residues. The result reported by Lundin et al. (17), with their very different reporter, fits perfectly into our length series.
The phenomenon of loop size affecting the reintegration efficiency of a downstream H segment is not unique to the DPAPB sequence used in our RI-DP constructs. We also tested a cytosolic loop sequence derived from carboxypeptidase Y (CPY; Prc1p) in the otherwise unchanged construct. With a total loop length of 145 residues, reintegration efficiency barely reached 50% at ∼8 leucines and did not improve with higher hydrophobicities (Fig. 1 G and H). Reducing the sequence to only 22 residues resulted in a reduction of the threshold to less than three leucines. Again, hydrophobicities of more than nine leucines even resulted in reduced reintegration efficiency.
Reintegration after a Generic Glycine-Serine Repeat or a Natural Intrinsically Disordered Sequence Is Efficient and Largely Length Independent.
Upon changing its length, the properties of the cytoplasmic spacer are altered simultaneously in different aspects: length affects its molecular mass, the time until the reintegration sequence emerges from the ribosome after stop-transfer integration, the maximal distance between the transmembrane segments, potential binding of chaperones, and the polypeptide conformation in general. Since the loop sequence is only a fragment of the DPAPB fold (consisting of one and a half blades of the eight-bladed propeller domain) or of the CPY structure (the propeptide and four nonconsecutive strands of a central β-sheet), it is unable to attain a stable native structure. It is expected to collapse to a molten globule (i.e., an ensemble of dynamic, compact, and partially folded conformations of a protein) with substantial secondary structure, little tertiary structure, and increased, solvent-exposed, and hydrophobic surface area, relative to the native state (22).
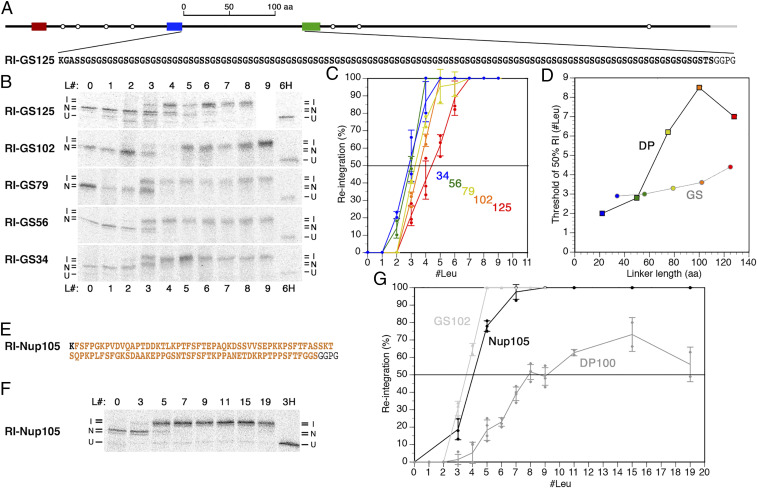
To reduce the complexity of the spacer sequence, we tested generic sequences of glycine-serine repeats of similar lengths (34 to 125 residues; Fig. 2A). These sequences are highly water soluble and flexible, cannot interact with chaperones, and, lacking hydrophobic residues, do not collapse on themselves. Reintegration efficiency was found to be high for all glycine-serine sequences with 50% thresholds between ∼2.9 to 3.6 leucines for lengths of 34 to 102 residues and ∼4.4 for 125 residues (Fig. 2 B and C). The length dependence of reintegration efficiency was thus much lower than for the DPAPB (or CPY) loops but still detectable (Fig. 2D).
Fig. 2.
Reintegration after a generic glycine-serine repeat or a natural intrinsically disordered sequence is efficient and largely length independent. (A) The construct RI-GS125 is schematically illustrated, and its generic glycine-serine repeat sequence between stop transfer and H segment is shown. (B) RI-GS125 and constructs with truncated GS repeat sequences (RI-GS102–34) were expressed, radioactively labeled, and analyzed, as in Fig. 1D. L# indicates the number of leucines in the H segment. Based on the glycosylation pattern, integration (I) or nonintegration (N) of the H segment, as well as uninserted unglycosylated products (U) were distinguished. Construct RI-GS125-L9 was not obtained, and the corresponding lane was thus replaced by white space. (C) The fraction of H segment reintegration was plotted versus the number of leucine residues in the H segment (mean, SD, and the individual values of at least three independent experiments). (D) The interpolated hydrophobicity threshold for 50% reintegration is plotted versus the linker length between stop transfer and H segment for the DP constructs (squares; from Fig. 1E) and the GS constructs (from C). (E) The sequence between stop transfer and H segment, containing residues 614 to 711 of Nup1p. (F) Autoradiographs of RI-Nup105 constructs with different H segments after radioactive labeling and gel electrophoresis. (G) The fraction of H segment reintegration was plotted versus the number of leucine residues in the H segment (mean, SD, and the individual values of at least three independent measurements).
Glycine-serine repeats are rather artificial and extremely flexible. We therefore also tested a natural intrinsically disordered sequence (i.e., a 100–amino acid segment) from the yeast nucleoporin Nup1 (residues 614 to 711), containing six FXF or FXFG motifs and forming relaxed or extended coils (23). The segment used has a complex amino acid composition, including a fair amount of apolar and charged residues (Fig. 2E). With this sequence as the cytoplasmic loop, reintegration was almost as efficient as glycine-serine repeats of the same length with an integration threshold of ∼4 leucines (Fig. 2 F and G).
Reintegration Is More Efficient after Rapidly Folding than after Folding-Deficient Sequences.
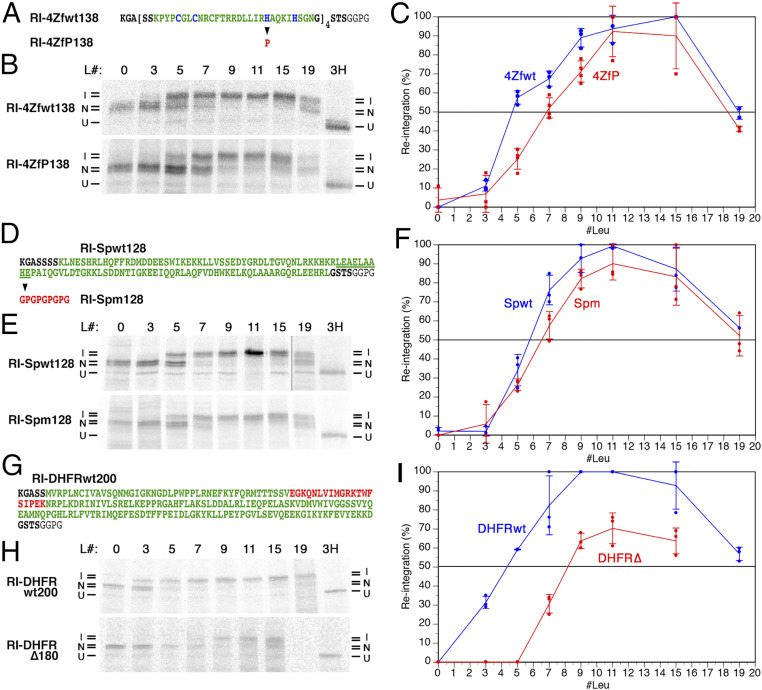
Contrasting disordered sequences, we also tested rapidly and stably folding domains on their effect on reintegration. For this, we cloned four successive wild-type (wt) zinc finger domains amounting to a total length of 138 residues in front of the reintegration H segments (RI-4ZFwt138; Fig. 3A). Zinc fingers have been demonstrated to cotranslationally fold while still in the ribosomal exit tunnel (24). In a second series, the first of two histidines coordinating with the zinc ion was mutated in all four domains to prevent native folding (RI-4ZfP138). Reintegration with wt domains showed a hydrophobicity threshold of ∼5 leucines and was thus clearly more efficient compared to mutant zinc fingers with ∼7 leucines (Fig. 3 B and C).
Fig. 3.
Effect of rapidly folding or mutated domains on the reintegration of a subsequent H segment. (A) Four consecutive copies of a zinc finger domain of yeast Adr1p (shown in green with the Zn2+-coordinating residues in blue) were inserted between the stop-transfer sequence and the reintegration H segment in construct RI-4Zfwt138. In RI-4ZfP138, one Zn2+-coordinating histidine was mutated to proline (in red) in each zinc finger to prevent folding. (B) Autoradiographs of the wt and mutant zinc finger constructs with different H segments are shown, as in Fig. 1D. L# indicates the number of leucines in the H segment. Based on the glycosylation pattern, integration (I) or nonintegration (N) of the H segment, as well as uninserted unglycosylated products (U) were distinguished. (C) The fraction of H segment reintegration was plotted versus the number of leucine residues in the H segment (mean, SD, and the individual values of at least three independent experiments). (D–F) Spectrin domain 16 of human spectrin α-chain (green) was inserted between the stop-transfer sequence and the reintegration H segment in construct RI-Spwt128. For RI-Spm128, nine underlined, central residues were replaced by a glycine/proline repeat sequence (red) to prevent native folding. Autoradiographs of the expressed proteins with different H segments were analyzed and the fraction of H segment reintegration quantified as above (B and C). (G–I) The wt DHFR was inserted between the stop-transfer sequence and the reintegration H segment in construct RI-DHFRwt200. In RI-DHFR∆180, the red sequence was deleted. Autoradiographs of the expressed proteins with different H segments were analyzed and the fraction of H segment reintegration quantified as above (B and C).
An alternative pair of wt and mutated sequences was based on a spectrin domain, a 105-residue, three-helix structure, that has been shown to rapidly fold as it emerges from the ribosomal exit cavity (25). To disturb native folding, a 9-residue central region of the middle helix was replaced by a disruptive glycine-proline repeat sequence (Fig. 3D). Again, the expression of reporter constructs with the wt (RI-Spwt128) and the mutant spectrin sequence (RI-Spm128) revealed an increase in hydrophobicity threshold upon mutation, if only by ∼1 leucine unit (Fig. 3 E and F).
A more dramatic difference in integration efficiency was observed between wt dihydrofolate reductase (DHFR), a globular enzyme of 187 residues, and a mutant version lacking a central portion of 20 residues (a β-strand and part of a helix in the folded structure), when inserted into the cytosolic loop preceding the RI–H segment (Fig. 3G). With wt DHFR, 50% reintegration was reached with ∼4 leucines and full reintegration with 9 and 11 (Fig. 3 H and I). With the mutant sequence, 50% reintegration was obtained only with ∼8 leucines, and full integration was not reached.
These results suggest that domain folding promotes reintegration compared to a very similar but folding-deficient mutant sequence. Interestingly, even with the rapidly folding domains, reintegration became inefficient at the highest H segment hydrophobicities tested (15 and 19 leucines).
Chaperone Binding to the Cytoplasmic Loop Facilitates Reintegration.
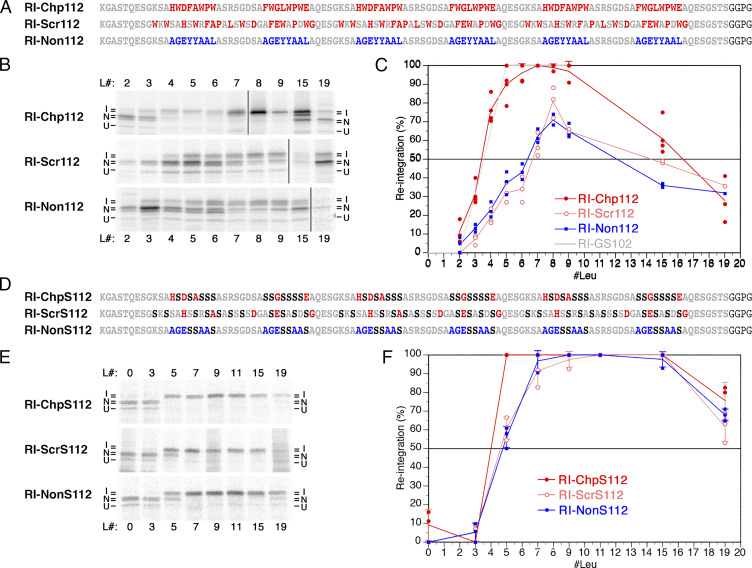
Slowly folding and folding-deficient proteins are potential substrates for molecular chaperones (26–28). By binding to exposed hydrophobic sequences, they increase solubility and reduce aggregation. To test a potential influence of chaperone binding to the cytosolic loop preceding the H segment, we employed sequences that have been identified experimentally by phage display to be recognized with high affinity by BiP (binding protein), a representative member of the Hsp70 family of chaperones in the ER (29). Hsp70 chaperones recognize segments of ∼7 residues enriched in hydrophobic amino acids (29, 30). In construct RI-Chp112, six chaperone-binding sequences were separated by hydrophilic peptides on a total loop length of 112 residues (Fig. 4A). As negative controls, the sequences were scrambled in RI-Scr112 or peptide sequences, including hydrophobic residues that had been demonstrated not to bind BiP (29), were inserted in RI-Non112.
Fig. 4.
Chaperone-binding sequences facilitate reintegration. (A) Three repeats of two different octapeptides (red) that have been shown to bind with high affinity to BiP (29), separated by hydrophilic octapeptides (gray), were inserted between the stop-transfer sequence and the reintegration H segment in construct RI-Chp112. In RI-Scr112, the sequence was scrambled to abolish chaperone binding. In RI-Non112, repeats of an octapeptide were used that had been shown not to bind BiP (blue) (29). (B) Autoradiographs of the above constructs with different H segments are shown, as in Fig. 1D. L# indicates the number of leucines in the H segment. Based on the glycosylation pattern, integration (I) or nonintegration (N) of the H segment, as well as uninserted unglycosylated products (U) were distinguished. (C) The fraction of H segment reintegration was plotted versus the number of leucine residues in the H segment (mean, SD, and the individual values of at least three independent experiments). For comparison, the tracing of the curve for RI-GS102 of the similar loop length from Fig. 2C is indicated in gray. (D–F) In constructs RI-ChpS112/ScrS112/NonS112, the hydrophobic residues (L, F, P, and W) in the segment between stop-transfer sequence and reintegration H segment were replaced by serines (in black; D). Upon expression and autoradiography (E), the fraction of H segment reintegration (F) was plotted, as above.
The chaperone-binding sequence allowed for efficient reintegration with a threshold of ∼3.5 leucines (Fig. 4 B and C), very much like that of the glycine-serine repeat sequence of comparable length, reaching 100% at six to nine leucines before declining again at very high hydrophobicities of 15 and 19 leucines. In contrast, the scrambled and the nonbinding sequences required, between six and seven leucines for 50% integration, did not reach complete reintegration with any H segment, similar to the long DPAPB loops. This result is consistent with chaperone binding keeping the polypeptide extended and soluble, whereas the scrambled and nonbinding sequences collapse on their hydrophobic residues.
Hydrophobic resides are also required for chaperone binding. Upon their replacement by serines in all three constructs (resulting in RI-ChpS112, RI-ScrS112, and RI-NonS112; Fig. 4D), we expected both chaperone binding and polypeptide collapse to be prevented. Consistent with this expectation, the mutated version of the chaperone-binding sequence remained very efficient for reintegration, while those of the scrambled and the nonbinding sequence improved their efficiencies for all H segments (Fig. 4 E and F).
Stop-Transfer Integration Efficiency Similarly Depends on the Folding State of the Upstream (Luminal) Sequence.
The above results raised the question whether membrane integration of a stop-transfer sequence is similarly affected by the preceding luminal sequence. Stop-transfer construct ST-DP128, where the initial signal anchor is separated by 128 residues of DPAPB from a stop-transfer H segment, was used as a starting point (Fig. 5A). As in previous studies (12, 13, 21), the transition of the fully translocated, sevenfold glycosylated product to the integrated, only fourfold glycosylated form occurred at slightly less than four leucines (Fig. 5 B and C). Truncation of the connecting sequence to 50 residues removed all but one potential glycosylation site, which turned out to be nonfunctional in the new context. As a result, stop transfer caused a shift from threefold glycosylated to unglycosylated products. The transition occurred already with H segments of one and two leucines. This shows that a substantial amount of protein unable to acquire a stable fold also reduces the integration efficiency of a downstream stop-transfer segment.
Fig. 5.
Stop-transfer integration similarly depends on the folding properties of the upstream sequence as reintegration. (A) Schematic representation of the original stop-transfer construct ST-DP128 with a DPAPB fragment of 128 residues separating the signal anchor (red) from the stop-transfer H segment (blue). In ST-DP50, the length of this spacer was truncated to 50 residues as indicated. In addition, constructs with a wt or a mutant spectrin domain (ST-SPwt161 and ST-Spm161, respectively; Fig. 3D), with a 100-residue glycine-serine repeat sequence (ST-GS144; Fig. 2A) and the chaperone-binding, -scrambled, or -nonbinding sequences (ST-Chp/Scr/Non144; Fig. 4A), were inserted into the luminal spacer after the second glycosylation site (pink line). The number in the names indicates the total lengths between stop transfer and H segment. (B–I) The indicated constructs were expressed, labeled with [35S]methionine, and analyzed by SDS–gel electrophoresis and autography (B, D, F, and H). L# indicates the number of leucines in the H segment. Based on the glycosylation pattern, integration (I) or translocation (T) of the H segment, as well as uninserted unglycosylated products (U) were distinguished. The fraction of H segment integration was plotted versus the number of leucine residues in the H segment (mean, SD, and the individual values of at least three independent experiments; C, E, G, and I). Stop-transfer integration was calculated as the twice-glycosylated (loop translocated) products, as a percentage of both loop-translocated and fully translocated products. For ST-DP50 (B), the single potential glycosylation site in the spacer was found not functional. Stop-transfer integration thus resulted in an increase of the unglycosylated fraction, which was thus used as a measure of integration. For ST-GS144 (F), an additional product with three glycans (*) was increasingly obtained with H segments containing ≥4 leucines. This form most likely corresponds to products that failed to translocate the luminal loop but instead translocated the C-terminal domain, as a result of the H segment acting as a reintegration sequence. Stop-transfer efficiency was determined as the fraction of the twice-glycosylated (loop translocated) products of the sum of proteins with two and five glycans, while reintegration (RI*; shown in gray) was determined as the percentage of threefold glycosylated products of the sum of proteins with three and zero glycans (G).
We further tested the effect of the rapidly folding spectrin domain and its mutant form, of intrinsically disordered glycine-serine repeat sequences and of chaperone-binding and -nonbinding sequences on stop-transfer efficiency, by inserting them into the translocated loop in a manner retaining the first two glycosylation sites (Fig. 5A, Bottom). The transition of fully glycosylated (C-translocated) to only loop glycosylated (integrated) forms occurred at ∼2.5 leucines for the wt spectrin domain and was shifted to ∼3.5 leucine upon mutation of the fold (Fig. 5 D and E). Interestingly, the wt spectrin domain resulted in a substantial amount of unglycosylated products not observed with the mutant domain. This may suggest that a fraction of the wt domain already folds in the cytosol, thus blocking translocation.
A high fraction of unglycosylated products was also detected for ST-GS144, in which the luminal domain consisted mostly of glycine-serine repeats (Fig. 5F). It has previously been reported that intrinsically disordered sequences are poorly translocated (31), which would also be expected for the GS repeats. The glycosylated fraction that was successful in initiating translocation changed from fully (fivefold) glycosylated, and thus translocated, to twice glycosylated (i.e., integrated) products with a threshold at ∼2 leucines (Fig. 5 F and G), indicative of efficient stop-transfer integration. However, with H segments containing four or more leucines, an additional threefold glycosylated product was generated, indicative of polypeptides with a cytosolic glycine-serine sequence and an H segment that had reinitiated translocation of the C-terminal sequence. The reintegration threshold was observed at five leucines, consistent with what one would expect for a glycine-serine sequence of that length (compared with Fig. 2C).
The effect of chaperone-binding and nonbinding sequences on the stop-transfer integration of a downstream H segment is particularly interesting, since it has been shown for posttranslational substrates that BiP binding acts as a molecular ratchet to achieve active translocation (32). One might have speculated that sequences with high affinity for BiP would favor translocation of the H segment at the expense of integration. However, we observed the opposite: a luminal loop containing multiple chaperone-binding peptides allowed the efficient integration of the following H segment with a threshold of ∼2 leucines, whereas a scrambled version of the same sequence or a sequence of nonbinding peptides showed reduced integration with thresholds of ∼4 and ∼5 leucines, respectively (Fig. 5 H and I). Chaperone binding thus favors integration equally for stop transfer and for reintegration.
Discussion
Topogenesis of membrane proteins is complex since several processes occur simultaneously. Translation is accompanied by the folding of the polypeptide as it emerges from the narrow parts of the ribosome tunnel or of the translocon. Within the translocon, the polypeptide has access to the lipid environment through the lateral gate and thus can equilibrate according to its properties. In addition, sequences looping out into the cytosol or into the ER lumen may interact with chaperones of the respective compartment. The interplay of these processes is still poorly understood.
Earlier models considered integration of transmembrane domains separately from folding of the hydrophilic portions of the protein. This made good sense, particularly when considering sequences passing through the translocon single file in a stop-transfer situation. The narrow conditions in the translocation pore isolate the segment inside from the preceding and the following sequences, offering the choice between the interior of the pore and the lipid environment. This is the basis for the concept of thermodynamic equilibration underlying the integration process. It yielded a very useful biological hydrophobicity scale readily applicable to calculate the integration propensity for any segment of polypeptide sequence. However, in different systems (model proteins, organisms, transmembrane orientations, etc.), apparent contributions to membrane integration varied considerably (summarized in ref. 1).
That cotranslational protein folding can affect translocation and integration, as has been observed previously. A zinc finger domain, which can fold already in the ribosomal exit tunnel and vestibule, was shown to block translocation across the translocon (33). Intrinsically disordered sequences were shown to be poorly translocated (31), and we made the same observation for glycine-serine sequences (Fig. 5 F and G). In addition, it has been observed that the integration efficiency of a stop-transfer sequence is markedly affected by up to ∼100 residues of the C-terminal sequence (21). The conformation of the nascent polypeptide in the space between ribosome and translocon considerably influenced the release of the H segment into the bilayer. Furthermore, known complications are closely spaced transmembrane domains that can only enter the translocon together (34) and specific interactions between transmembrane domains (35). Orientational preferences of successive transmembrane domains enhance integration of otherwise insufficiently hydrophobic segments (36). To avoid these issues, our model constructs typically present H segments clearly spaced from other transmembrane domains.
In the present study, we discovered that the sequence upstream of the H segment can have a large effect on membrane integration. Effects were found similarly for stop transfer and for reintegration. High-integration efficiency was observed, when the preceding sequence was short or—if it was substantially longer—when it was highly soluble and flexible, as is the case for generic glycine-serine repeats or for natural intrinsically disordered sequences. Such sequences had low-hydrophobicity thresholds for 50% integration in the range of two to four leucines for reintegration and ∼2 leucines for stop transfer. This is in contrast to longer fragments of DPAP or CPY that required up to ∼8 leucines for reintegration and ∼4 for stop transfer. The high-integration thresholds for longer protein fragments appear to be connected to their inability to acquire a stable fold. This is supported by the finding that, when rapidly folding protein sequences (zinc finger, spectrin domain, and DHFR) were inserted before the H segment, the integration thresholds were always increased upon mutation. The effect of the mutations differed in extent (from +1 to +4 leucines), probably reflecting the extent to which the domains were disrupted by the respective mutations.
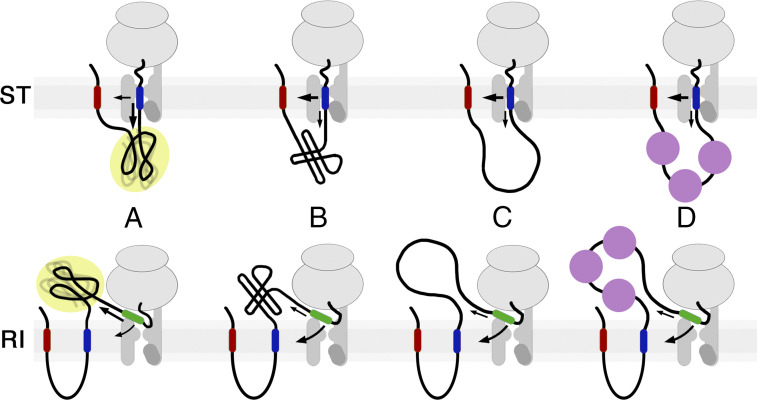
When a nascent polypeptide emerges from the ribosome, it compacts vectorially. Emerging segments acquire secondary structures (in particular helices) that dock onto one another and continue to rearrange, while conformationally fluctuating, until the sequence is complete to reach the native state (24, 25, 37). Mutations or deletions that prevent final domain stabilization keep conformations dynamic with the substantial exposure of hydrophobic surfaces. We propose that such partially folded polypeptides offer hydrophobic surfaces as an alternative environment for potential transmembrane domains in competition with the translocon and the membrane (schematically illustrated in Fig. 6), thereby reducing the efficiency of membrane integration and requiring higher H segment hydrophobicity. Accordingly, DPAPB fragments of increasing length provide increasing hydrophobic surface for the H segment to interact with, resulting in decreasing integration efficiency. This phenomenon is in agreement with the observation by Kuroiwa et al. (38) in in vitro experiments that reintegration efficiency of an L10 sequence was reduced from ∼65% to ∼50% and ∼30% upon increasing the preceding loop length from 58 to 100 and 200 residues, respectively.
Fig. 6.
How the folding state of upstream sequences affects membrane integration. A nascent chain emerging from the ribosome (in the case of reintegration; RI) or from the luminal side of the translocon (in the case of stop transfer; ST) will compact vectorially, acquire some secondary and tertiary structure but remain conformationally fluctuating, and expose substantial hydrophobic surfaces that—if the sequence cannot reach a native fold (as for protein fragments or mutant domains)—will compete with translocon/membrane for the transmembrane segment and affect integration efficiency (A). If the sequence is folded rapidly (B), if it is intrinsically disordered (C), or if the sequence is bound by chaperones (D), they interact little with potential transmembrane domains, thus allowing efficient integration with low-hydrophobicity thresholds.
In contrast, folded domains that present hydrophilic surfaces are rather inert toward subsequent hydrophobic segments, allowing for efficient H segment integration. This is also the case for sequences without hydrophobic residues like glycine-serine repeats and for natural intrinsically disordered polypeptides that have evolved to remain unfolded in solution without collapsing on their apolar residues.
For reintegration, the partially folded upstream sequence directly competes with the translocon and the membrane for the emerging H segment. For stop transfer, however, an H segment first encounters the membrane from within the translocon before it may interact with the already translocated upstream sequence. The observed effects suggest that translation and translocation are not tightly coupled, allowing H segments to reversibly reach into the ER lumen to experience the environment of the nonnative fold of the preceding polypeptide. This is less the case with higher H segment hydrophobicity that more efficiently directs it into the bilayer, limiting luminal exposure. This explains the smaller range of thresholds observed for stop transfer than for reintegration (two to four leucines and three to seven leucines, respectively, for DP50 to DP128 segments) and why for stop transfer, unlike for reintegration, complete integration is always obtained with higher H segment hydrophobicities.
Molecular chaperones, in particular Hsp70 family members, have been shown to associate with nascent chains emerging from the ribosome into the cytosol to keep them in a folding, competent state and prevent aggregation (39). They are also known to bind to chains appearing on the other side of the membrane to drive as a ratchet (posttranslational) translocation of proteins into the ER or mitochondria (32, 40). Hsp70 chaperones bind short sequences of ∼7 residues containing several hydrophobic amino acids (41) that will be hidden inside the protein as soon as it is folded but that are not apolar or long enough to integrate into the membrane. The peptide-binding configuration of Hsp70 proteins is evolutionarily conserved, with some variability in the register and orientation of bound peptides, providing plasticity and promiscuity in substrate binding (28). Kar2p (BiP) is the main yeast chaperone in the ER lumen with respect to newly synthesized proteins. The major cytosolic Hsp70 proteins to catalyze protein folding are Ssa1–4p and the ribosome-associated Ssb1/2p.
We actually expected strong chaperone-binding sequences in the cytosolic or luminal segment to reduce membrane insertion of a following H segment by pulling it away from the translocon by Brownian motion. The results revealed the opposite effect: increased membrane integration for chaperone-binding sequences with a threshold of three to four leucines versus six to seven with scrambled or nonbinding sequences for reintegration and a threshold of ∼2 versus 4 to 5 leucines, respectively, for stop transfer. Indeed, the results are consistent with our model: chaperone binding protects the polypeptide from hydrophobic collapse and thus from forming a dynamic assembly of hydrophobic surfaces that compete for the adjacent H segment. It also suggests that the natural DPAPB or CPY fragments or the mutant sequences that we tested were not comparable in chaperone binding to the high-affinity peptide sequences we have used.
For most reintegration constructs, we observed that the reintegration efficiency decreased for maximally hydrophobic H segments of 19 leucines. It suggested that H segments can be too hydrophobic. This phenomenon has already been observed in in vitro experiments with sequences of 5 to 15 consecutive leucines (38): reintegration was reduced with L15 sequences. We can only speculate whether long oligo-leucine segments in part bind flat onto the lipid membrane in a nonproductive manner or the formation of a hairpin with a hydrophilic downstream sequence to be inserted into the translocon is energetically disfavored.
Overall, our study shows that the integration of potential transmembrane segments is not independent of the rest of the polypeptide but that the conformational properties of the preceding polypeptide may interfere with the well-established equilibration process. The results suggest that rapid folding of cytoplasmic and luminal domains is a prerequisite for efficient and complete transmembrane integration.
Materials and Methods
Yeast Strain and Model Proteins.
Yeast strain RSY1293 (mata, ura3-1, leu2-3,-112, his3-11,15, trp1-1, ade2-1, can1-100, sec61::HIS3, and [pDQ1]) was used (42). The coding sequence of DPAPB (DAP2) was used to construct model proteins, as illustrated in Fig. 1A, inserting different H segments as introduced by Hessa et al. (10). Sequences from CPY (PRC1), a zinc finger of yeast Adr1p (residues 130 to 158), spectrin domain 16 of human spectrin α chain (residues 1,759 to 1,870), mouse DHFR, a segment of nucleoporin Nup1p (residues 614 to 711), and artificial sequences of glycine-serine repeats or composed of chaperone-binding, scrambled, or nonbinding sequences were inserted into the model proteins, replacing the loop sequences upstream of the H segments. Constructs were made by PCR to introduce restriction sites for fragment exchange or using annealed, synthetic complementary oligonucleotides with appropriate protruding ends. Protein sequences of the model proteins analyzed in this study are provided in Figs. 1–4 and SI Appendix, Tables S1 and S2. Constructs were made in the expression plasmid pRS426 (URA3 2µ) with a glyceraldehyde-3-phosphate dehydrogenase promoter.
Labeling and Immunoprecipitation.
Yeast transformants were grown overnight in synthetic defined (SD) medium lacking uracil at 30 °C, diluted to OD600 0.6, and regrown for 2 h. A total of 1.5 mL cell suspension was harvested, resuspended in 150 µL SD lacking uracil and methionine, incubated for 15 min at 30 °C, and pulse labeled for 7 min with 150 µCi/mL [35S] methionine/cysteine (PerkinElmer) at 30 °C. At 4 °C, translation was stopped by adding 2.5 μL 1-M NaN3. Cells were pelleted; washed with 150 μL phosphate-buffered saline; resuspended in 150 μL 50-mM Tris⋅HCl, pH 7.5, 5 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride; lysed with 100-µL glass beads for 10 min in a bead beater; supplemented with 1% SDS; and heated to 95 °C for 5 min. Cell remnants were removed by centrifugation for 10 min in a microfuge, the supernatant was immunoprecipitated overnight at 4 °C with mouse monoclonal anti-HA antibodies (HA.11, clone 16B12; 1:1,000) in 800 µL TNET (30 mM Tris⋅HCl, pH 7.5, 120 mM NaCl, 5 mM EDTA, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor mixture) and for 1 h with Sepharose A-beads, washed with TNET, and analyzed by SDS–gel electrophoresis and autoradiography. Signals were quantified using a phosphorimager. Each replicate was performed using cells from separate transformations. For deglycosylation, the samples boiled in gel–sample buffer were incubated for 1 h with 1 mU endo-β-d-N-acetyl glucosaminidase H for 1 h at 37 °C before gel electrophoresis.
Supplementary Material
Acknowledgments
We thank Dr. Peter Pohl, Johannes Kepler University Linz, and Dr. Sabine Rospert, University of Freiburg, for their support. This work was supported by Grant 31003A-182519 from the Swiss National Science Foundation (to M.S.) and by Grant W1250 from the Austrian Science Fund (to M.Z.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102675118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Spiess M., Junne T., Janoschke M., Membrane protein integration and topogenesis at the ER. Protein J. 38, 306–316 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Rapoport T. A., Li L., Park E., Structural and mechanistic insights into protein translocation. Annu. Rev. Cell Dev. Biol. 33, 369–390 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Voorhees R. M., Fernández I. S., Scheres S. H. W., Hegde R. S., Structure of the mammalian ribosome-Sec61 complex to 3.4 Å resolution. Cell 157, 1632–1643 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gogala M., et al., Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion. Nature 506, 107–110 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Voorhees R. M., Hegde R. S., Structure of the Sec61 channel opened by a signal sequence. Science 351, 88–91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Heijne G., The signal peptide. J. Membr. Biol. 115, 195–201 (1990). [DOI] [PubMed] [Google Scholar]
- 7.Beltzer J. P., et al., Charged residues are major determinants of the transmembrane orientation of a signal-anchor sequence. J. Biol. Chem. 266, 973–978 (1991). [PubMed] [Google Scholar]
- 8.Parks G. D., Lamb R. A., Topology of eukaryotic type II membrane proteins: Importance of N-terminal positively charged residues flanking the hydrophobic domain. Cell 64, 777–787 (1991). [DOI] [PubMed] [Google Scholar]
- 9.Denzer A. J., Nabholz C. E., Spiess M., Transmembrane orientation of signal-anchor proteins is affected by the folding state but not the size of the N-terminal domain. EMBO J. 14, 6311–6317 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hessa T., et al., Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433, 377–381 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Hessa T., et al., Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450, 1026–1030 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Junne T., Kocik L., Spiess M., The hydrophobic core of the Sec61 translocon defines the hydrophobicity threshold for membrane integration. Mol. Biol. Cell 21, 1662–1670 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demirci E., Junne T., Baday S., Bernèche S., Spiess M., Functional asymmetry within the Sec61p translocon. Proc. Natl. Acad. Sci. U.S.A. 110, 18856–18861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hessa T., Reithinger J. H., von Heijne G., Kim H., Analysis of transmembrane helix integration in the endoplasmic reticulum in S. cerevisiae. J. Mol. Biol. 386, 1222–1228 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Xie K., et al., Features of transmembrane segments that promote the lateral release from the translocase into the lipid phase. Biochemistry 46, 15153–15161 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Ojemalm K., Botelho S. C., Stüdle C., von Heijne G., Quantitative analysis of SecYEG-mediated insertion of transmembrane α-helices into the bacterial inner membrane. J. Mol. Biol. 425, 2813–2822 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Lundin C., Kim H., Nilsson I., White S. H., von Heijne G., Molecular code for protein insertion in the endoplasmic reticulum membrane is similar for N(in)-C(out) and N(out)-C(in) transmembrane helices. Proc. Natl. Acad. Sci. U.S.A. 105, 15702–15707 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Botelho S. C., et al., TIM23-mediated insertion of transmembrane α-helices into the mitochondrial inner membrane. EMBO J. 30, 1003–1011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsirigos K. D., Peters C., Shu N., Käll L., Elofsson A., The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 43, W401–W407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sommer N., Junne T., Kalies K.-U., Spiess M., Hartmann E., TRAP assists membrane protein topogenesis at the mammalian ER membrane. Biochim. Biophys. Acta 1833, 3104–3111 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Junne T., Spiess M., Integration of transmembrane domains is regulated by their downstream sequences. J. Cell Sci. 130, 372–381 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Fink A. L., Compact intermediate states in protein folding. Annu. Rev. Biophys. Biomol. Struct. 24, 495–522 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Yamada J., et al., A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Mol. Cell. Proteomics 9, 2205–2224 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nilsson O. B., et al., Cotranslational protein folding inside the ribosome exit tunnel. Cell Rep. 12, 1533–1540 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsson O. B., et al., Cotranslational folding of spectrin domains via partially structured states. Nat. Struct. Mol. Biol. 24, 221–225 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Hartl F. U., Bracher A., Hayer-Hartl M., Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Balchin D., Hayer-Hartl M., Hartl F. U., In vivo aspects of protein folding and quality control. Science 353, aac4354 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Rosenzweig R., Nillegoda N. B., Mayer M. P., Bukau B., The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 20, 665–680 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Blond-Elguindi S., et al., Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell 75, 717–728 (1993). [DOI] [PubMed] [Google Scholar]
- 30.Rüdiger S., Germeroth L., Schneider-Mergener J., Bukau B., Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 16, 1501–1507 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonsberg A., et al., The Sec61/SecY complex is inherently deficient in translocating intrinsically disordered proteins. J. Biol. Chem. 292, 21383–21396 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matlack K. E., Misselwitz B., Plath K., Rapoport T. A., BiP acts as a molecular ratchet during posttranslational transport of prepro-α factor across the ER membrane. Cell 97, 553–564 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Conti B. J., Elferich J., Yang Z., Shinde U., Skach W. R., Cotranslational folding inhibits translocation from within the ribosome-Sec61 translocon complex. Nat. Struct. Mol. Biol. 21, 228–235 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hedin L. E., et al., Membrane insertion of marginally hydrophobic transmembrane helices depends on sequence context. J. Mol. Biol. 396, 221–229 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Tu L., Khanna P., Deutsch C., Transmembrane segments form tertiary hairpins in the folding vestibule of the ribosome. J. Mol. Biol. 426, 185–198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ojemalm K., Halling K. K., Nilsson I., von Heijne G., Orientational preferences of neighboring helices can drive ER insertion of a marginally hydrophobic transmembrane helix. Mol. Cell 45, 529–540 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liutkute M., Maiti M., Samatova E., Enderlein J., Rodnina M. V., Gradual compaction of the nascent peptide during cotranslational folding on the ribosome. eLife 9, e60895 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuroiwa T., Sakaguchi M., Omura T., Mihara K., Reinitiation of protein translocation across the endoplasmic reticulum membrane for the topogenesis of multispanning membrane proteins. J. Biol. Chem. 271, 6423–6428 (1996). [DOI] [PubMed] [Google Scholar]
- 39.Kramer G., Shiber A., Bukau B., Mechanisms of cotranslational maturation of newly synthesized proteins. Annu. Rev. Biochem. 88, 337–364 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Okamoto K., et al., The protein import motor of mitochondria: A targeted molecular ratchet driving unfolding and translocation. EMBO J. 21, 3659–3671 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clerico E. M., Tilitsky J. M., Meng W., Gierasch L. M., How hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J. Mol. Biol. 427, 1575–1588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pilon M., Schekman R., Römisch K., Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 16, 4540–4548 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cymer F., von Heijne G., White S. H., Mechanisms of integral membrane protein insertion and folding. J. Mol. Biol. 427, 999–1022 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.