Fig. 2.
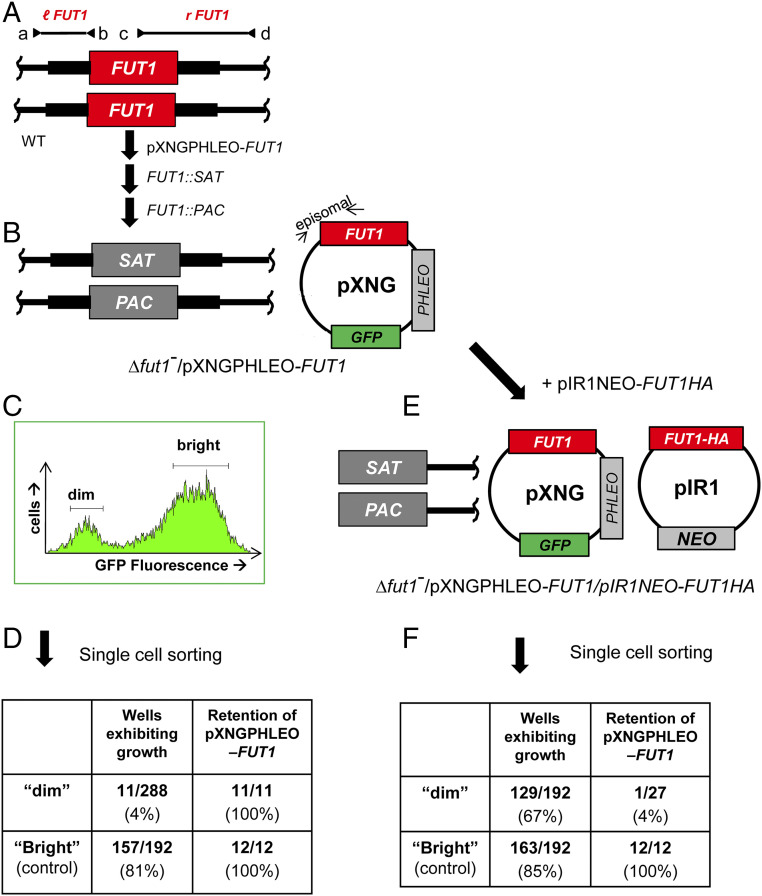
Plasmid segregation tests show FUT1 is essential. (A) FUT1 locus and transfection. The figure shows the WT FUT1 locus, with the ORF as a central red box and the flanking sequences used for homologous replacement by SAT or PAC drug resistance cassettes (FUT1::SAT or FUT1::PAC, respectively) shown as heavier flanking lines. The location of “outside” primers used to establish planned replacements (a/b or c/d; to confirm replacements SAT or PAC specific primers were substituted for b or c; SI Appendix, Fig. S3) are shown. WT L. major was first transfected with pXNGPHLEO-FUT1, and successively with the FUT1::SAT and FUT::PAC constructs. (B) The chromosomal null Δfut1− mutant obtained in the presence of episomal FUT1. The genotype of this line is FUT::∆SAT/FUT1::∆PAC/ +pXNGPHLEO-FUT1, abbreviated as Δfut1−/+pXNGPHLEO-FUT1. PCR confirmation tests of the predicted genotype are found in SI Appendix, Fig. S3. pXNG additionally expresses GFP (45). (C) Plasmid segregation tests of FUT1 essentiality. Δfut1−/+pXNGPHLEO-FUT1 was grown for 24 h in the absence of phleomycin, and analyzed by GFP flow cytometry. The two gates used for subsequent quantitation and/or sorting of parasites are shown; dim or weakly fluorescent (2–10 FU) and bright or strongly fluorescent (100–1,000 FU). (D) Results of sorting experiment shown in C. Single cells from both GFP dim or bright populations sorted into 96-well plates containing M199 medium, and incubated for 2 wk, at which time growth was assessed. The numbers sorted and their growth and properties are shown, with bright cells mostly surviving and dim cells mostly not. All 11 survivors from the dim and 12 tested from the bright population sort were confirmed to retain of pXNGPHLEO-FUT1 by growth in media containing phleomycin. (E) Generation of line required for plasmid shuffling. Δfut1−/+pXNGPHLEO-FUT1 was transfected with pIR1NEO-FUT1-HA expressing C-terminally tagged FUT1. PCR tests confirming the predicted genotype Δfut1−/+pXNGPHLEO-FUT1/+pIR1NEO-FUT1-HA are shown in SI Appendix, Fig. S3. This line was then grown 24 h in the absence of phleomycin and subjected to cell sorting and growth tests as described in C. (F) Results of sorting experiment described in E. In this experiment, both dim and bright cells mostly survived (67% and 85%, respectively). Twenty-seven of the dim cells were tested, of which 26 had lost pXNGPHLEO-FUT1 by PCR tests, thereby representing the desired Δfut1−/+pIR1NEO-FUT1-HA.