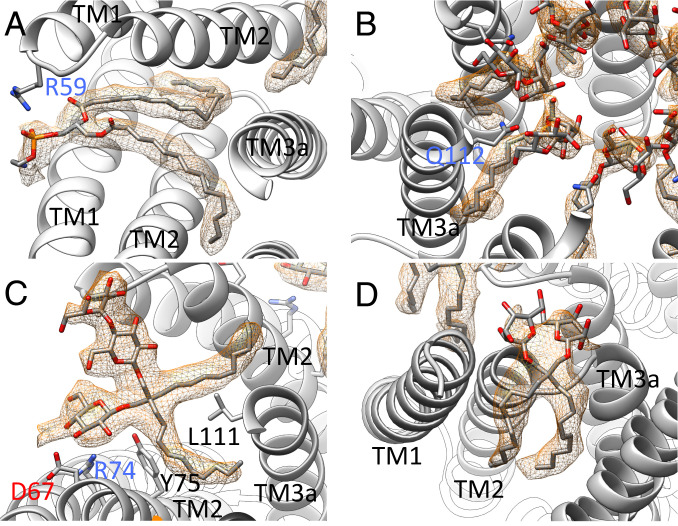
Fig. 4.
Selected ligands in the structure of open MscS in LMNG. (A) One lipid, modeled as DOPE, is resolved in the structure at the paddle loop. A salt bridge is formed with R59. The fatty acid chains are bent because the TM3a helices restricting the space in the open conformation (B) DDM molecules are located in the pore entering the gaps between the TM3a helices. The headgroup is coordinated by Q112. (C) An LMNG molecule close to the paddle loop and the lipid. This LMNG molecule, together with a second, cover a large hydrophobic opening toward the cytosol. One maltose unit is well resolved, while in the second unit, only one sugar ring is resolved. (D) One LMNG molecule per subunit of the LMNG stack on the periplasmic side. This LMNG is close to the TM2–TM3a loop and relates to a well-resolved lipid at this loop in the closed structure. The headgroups are not well resolved, and only one sugar ring each is shown.