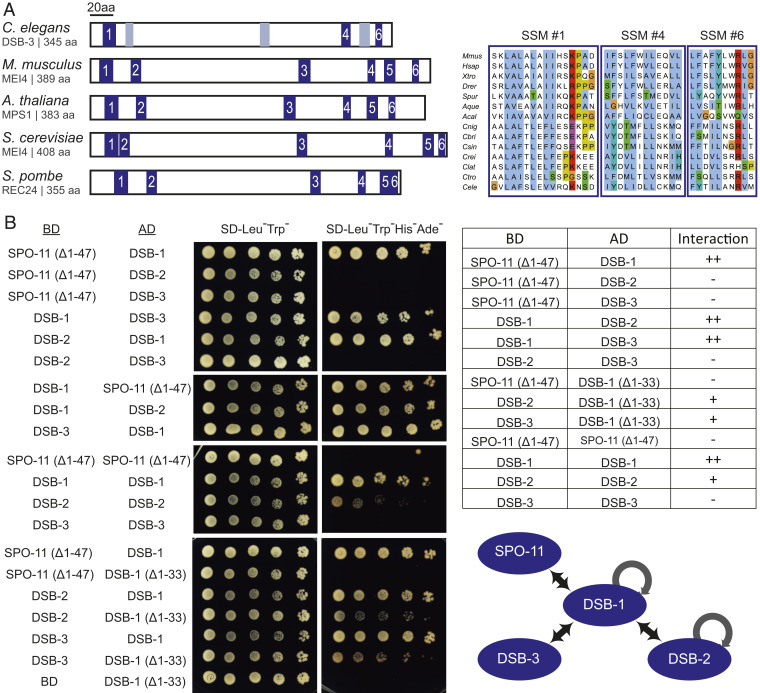
Fig. 4.
Evidence that DSB-1, DSB-2, and DSB-3 form a complex homologous to the yeast and mammalian REC114-MEI4 complexes. (A, Left) Schematic depicting the positions of six SSMs (blue boxes) previously defined for Mei4 homologs in diverse species, indicating the three SSMs (1, 4, 6) that are most strongly supported in C. elegans DSB-3; gray boxes indicate positions that potentially correspond to SSMs 2, 3, and 5 based on a multiple sequence alignment (MSA) with vertebrate and marine invertebrate homologs of similar size, but are less well conserved. (A, Right) Aligned sequences of SSMs 1, 4, and 6 from M. musculus, Homo sapiens, Xenopus tropicalis, Danio rerio, Strongylocentrotus purpuratus, Amphimedon queenslandica, Aplysia californica, and the following nematodes of the genus Caenorhabditis: C. nigoni, C. briggsae, C. sinica, C. remanei, C. latens, C. tropicalis, and C. elegans. SSMs are cropped from an MSA generated in JalView (78) using the ClustalX coloring scheme. For additional information, see SI Appendix, Fig. S2 and Supplementary Materials and Methods. (B) Y2H assay revealing interactions among the DSB-1, -2, and -3 proteins and between DSB-1 and SPO-11. Potential interactions between proteins fused with the GAL4 activation domain (AD) and proteins fused with the GAL4 DNA binding domain (BD) were assayed by growth on media lacking histidine and adenine. A construct producing an N-terminal truncation of SPO-11 lacking the first 47 amino acids (Δ1–47) was used for these analyses, as severe auto-activation was observed for full-length SPO-11. Negative controls showing lack of auto-activation for the constructs used are presented in SI Appendix, Fig. S2. A schematic summarizing the identified interactions is shown on the bottom right.