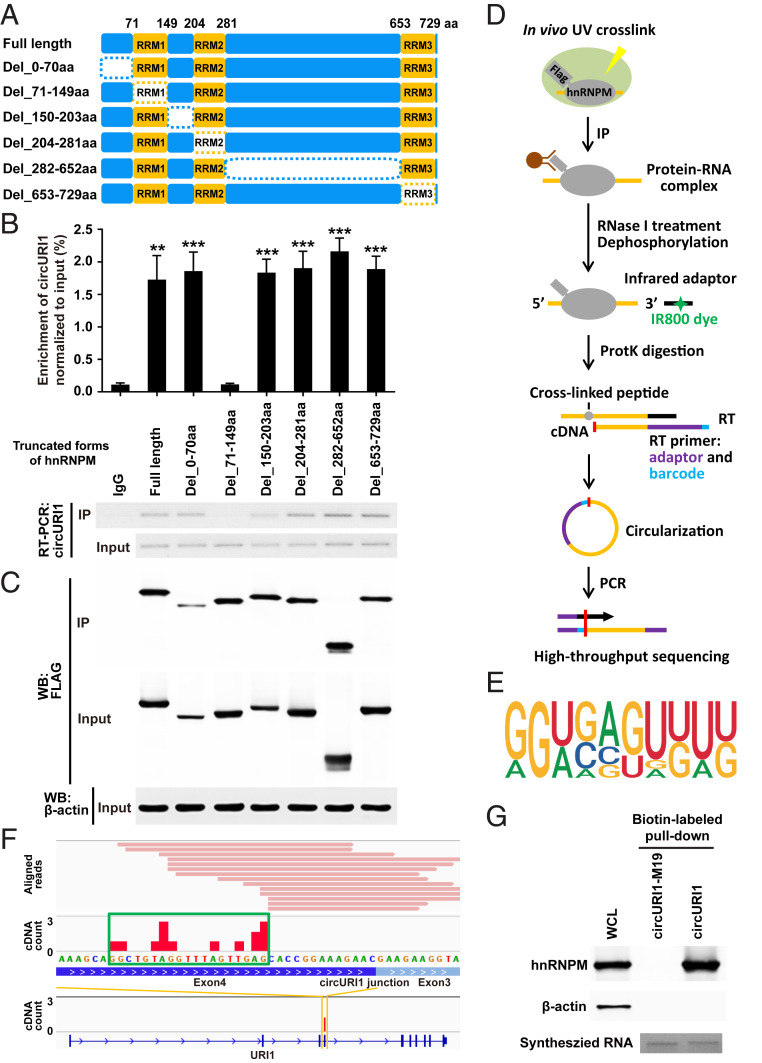
Fig. 4.
The RRM1 domain of hnRNPM and the 19-nt sequence of circURI1 are required for their interaction. (A) Schematic diagram revealing full-length hnRNPM and a variety of truncated forms of hnRNPM. The dotted rectangles indicate the truncated domains. (B and C) Association of circURI1 examined by RT-PCR and RT-qPCR with RNA IP of Flag-tagged full-length and truncated hnRNPM in AGS cells. Full-length and truncated forms of hnRNPM were determined by Western blot with an anti-FLAG antibody. β-actin was a loading control for Western blot. (D) Illustration of the experimental procedure of iCLIP-seq. (E) The binding motif of hnRNPM derived from iCLIP analyzed by Hypergeometric Optimization of Motif EnRichment (HOMER). (F) Individual Flag-hnRNPM iCLIP-seq reads (in pink) were aligned to circURI1 or URI1 mRNA. iCLIP-seq cDNA counts (in red) showing the binding signals. The binding site of hnRNPM on circURI1 deduced from iCLIP-seq is depicted with the green rectangle. (G) RNA pulldown assay of biotin-labeled circURI1 and circURI1 with the mutation of the 19-nt binding site (circURI1-M19) followed by Western blot with an anti-hnRNPM antibody. CircURI1 and circURI1-M19 were circularized in vitro. Agarose gel performed in vitro synthesized biotin-labeled circURI1 and circURI1-M19. β-actin was a negative control for Western blot. WCL, whole-cell lysis. Error bars indicate SEM from three independent experiments. **P < 0.01; ***P < 0.001 by two-tailed Student’s t test.