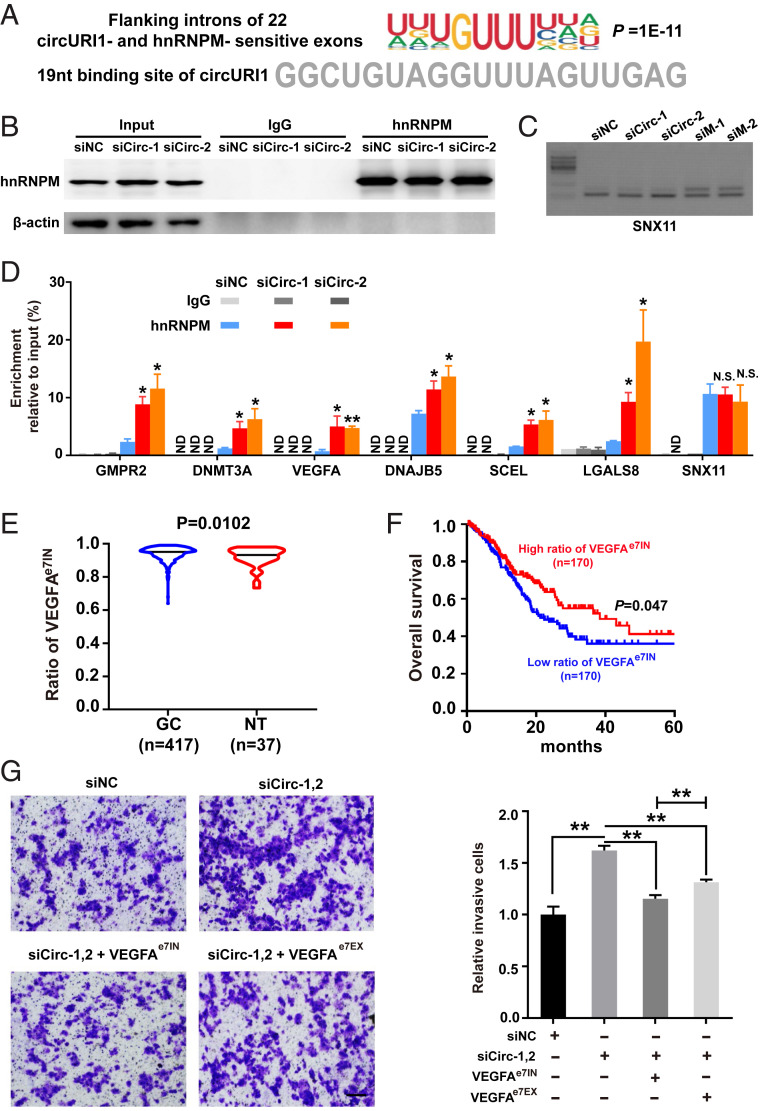
Fig. 7.
CircURI1 modulates alternative splicing via sequestering hnRNPM. (A) The deduced motif of introns flanked by 22 both circURI1 and hnRNPM-sensitive exons analyzed by HOMER (-len 10). The 19-nt sequence of circURI1 required for the interaction of hnRNPM is indicated below. (B) RNA IP efficiency of hnRNPM demonstrated by Western blot after circURI1 silencing in AGS cells. β-actin was a negative control for RNA IP. siNC, siRNA with scrambled sequences; siCirc-1 and siCirc-2, two siRNAs against the junction sites of circURI1. (C) Semi-quantitative RT-PCR validation of alternatively spliced exon in sorting nexin 11 (SNX11), a hnRNPM-sensitive gene not sensitive to circURI1, upon silencing circURI1 or hnRNPM in AGS cells. siM-1 and siM-2, two independent siRNAs against hnRNPM. (D) RNA IP of hnRNPM showing the binding for the corresponding pre-mRNAs of circURI1-sensitive targets upon circURI1 knockdown in AGS cells. SNX11 was a negative control. ND, no detection. (E) Violin plots depicting the ratio of VEGFAe7IN in GC and normal patients analyzed from TCGA database. NT, normal tissue. VEGFAe7IN, exon 7 inclusion of VEGFA. (F) Kaplan–Meier analysis of overall survival for GC patients with VEGFAe7IN from TCGA database. The red curve represents survival in GC patients with a high ratio of VEGFAe7IN, and the blue line represents survival in patients with a low ratio of VEGFAe7IN. (G) Transwell assays of AGS cells treated with siRNAs targeting circURI1 or co-overexpressing VEGFAe7IN or VEGFAe7EX. siNC, siRNA with scrambled sequences; siCirc-1,2, two siRNAs (siCirc-1, siCirc-2) against the junction sites of circURI1. (Scale bars, 100 μm.) Error bars indicate SEM from three independent experiments. In D, E, and G, N.S., not significant; *P < 0.05; **P < 0.01 by two-tailed Student’s t test. In F, P value by the log-rank test.