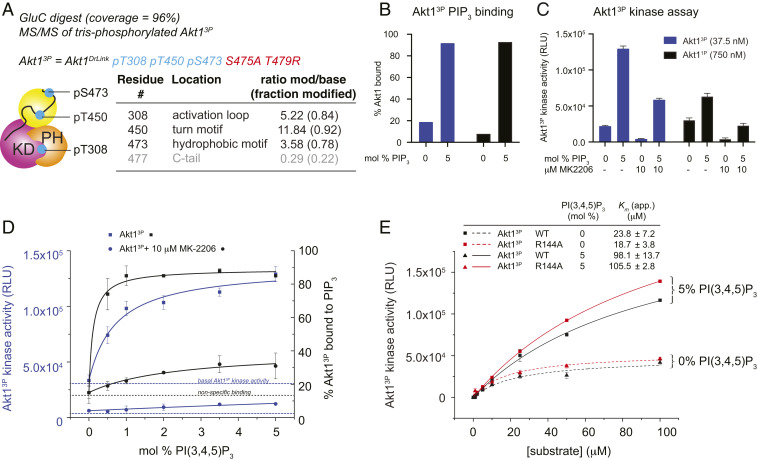
Fig. 3.
Phosphorylation does not override the requirement for PIP3. (A) Phosphorylation state analysis of Akt13P after coculture with A-443654 and okadaic acid. Tandem mass spectrometry of GluC digest. Additional substoichiometric phosphorylation of S477 in 4P species indicated in red. (B) Liposome pelleting assay for 0% and 5% PIP3 liposomes indicating the binding of Akt13P and Akt11P to PIP3 in the kinase assay shown in F. (C) Akt1 kinase assay ± PIP3 liposomes, ± 10 μM MK-2206. Akt13P (37.5 nM), black bars; Akt11P (750 nM), gray bars. (D) Kinase assay of Akt13P in the presence of liposomes containing increasing concentrations of PIP3. Left axis, and blue lines correspond to kinase activity. Right axis and black lines correspond to % PIP3 binding (determined by a liposome pelleting assay). Squares, Akt13P; circles, Akt13P preincubated for 10 min with 10 μM MK-2206. PIP3 binding and PIP3-dependent increase in kinase activity were fit to one-site binding models, taking into account basal Akt13P activity and nonspecific binding in the presence of 0 mol % PIP3 liposomes respectively. (E) Kinase assay of Akt13P and Akt13P R144A with increasing substrate concentration in the presence of liposomes containing 0 or 5 mol% PIP3. Error bars indicate the SD of three independent measurements.