FIGURE 3.
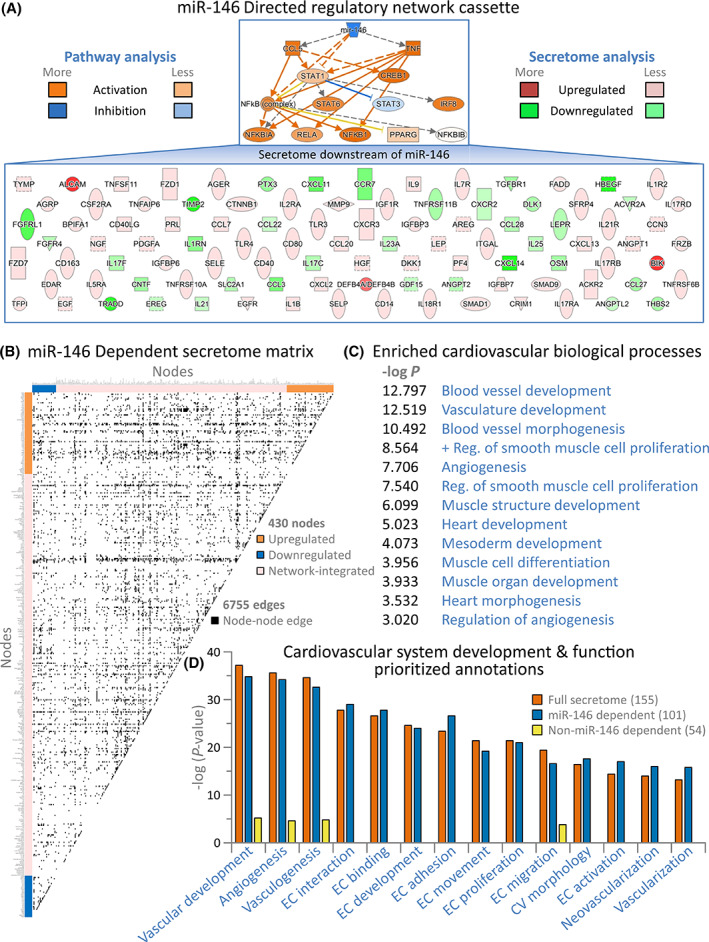
Secretome functionality concentrated within the miR‐146 dependent sub‐secretome. (A) Pathway interrogation pinpointed miR‐146 at the apex of a 14‐protein network upstream of the secretome (A, upper) comprising interdependent transcriptional regulators (TNFα; CREB1; PPARγ; CCL5; IRF8; STAT1; STAT3; STAT6; and the NFκB complex, including NFκB1, NFκBIA, NFκBIB, and RELA) predicted to be activated or inhibited (orange or blue, respectively). Each node of the miR‐146 directed network regulates one or more downstream components of a cardiopoietic sub‐secretome (A, lower), comprising 65% of the overall differential secretome (101 of 155 proteins), with protein increase or decrease shown (red or green, respectively). B, Integrating the 101 protein miR‐146 dependent sub‐secretome into an expanded interaction neighborhood generated a 430 node network, clustered along matrix axes by inclusion basis (colored bars), connected by 6755 interactions (edges) shown as black squares. Network node names and topological parameters are listed in supporting Table S3. C, BiNGO analysis of the miR‐146 dependent secretome network yielded 558 significantly enriched terms (P < .001; with the top 100 listed in supporting Table S4), and revealed enriched cardiovascular biological processes related to vascular, angiogenic, and smooth/cardiac muscle development (Positive regulation = + Reg.). D, Enriched cardiovascular system development and functions of the full secretome (orange) and the miR‐146 dependent (blue) and independent (yellow) sub‐secretomes indicated that proteins responsible for pro‐cardiovasculogenic functionality were largely contained within the miR‐146 dependent subset. Indeed, this sub‐secretome included 77%‐100% of proteins associated with individual enriched cardiovascular functions within the full secretome
