Figure 3.
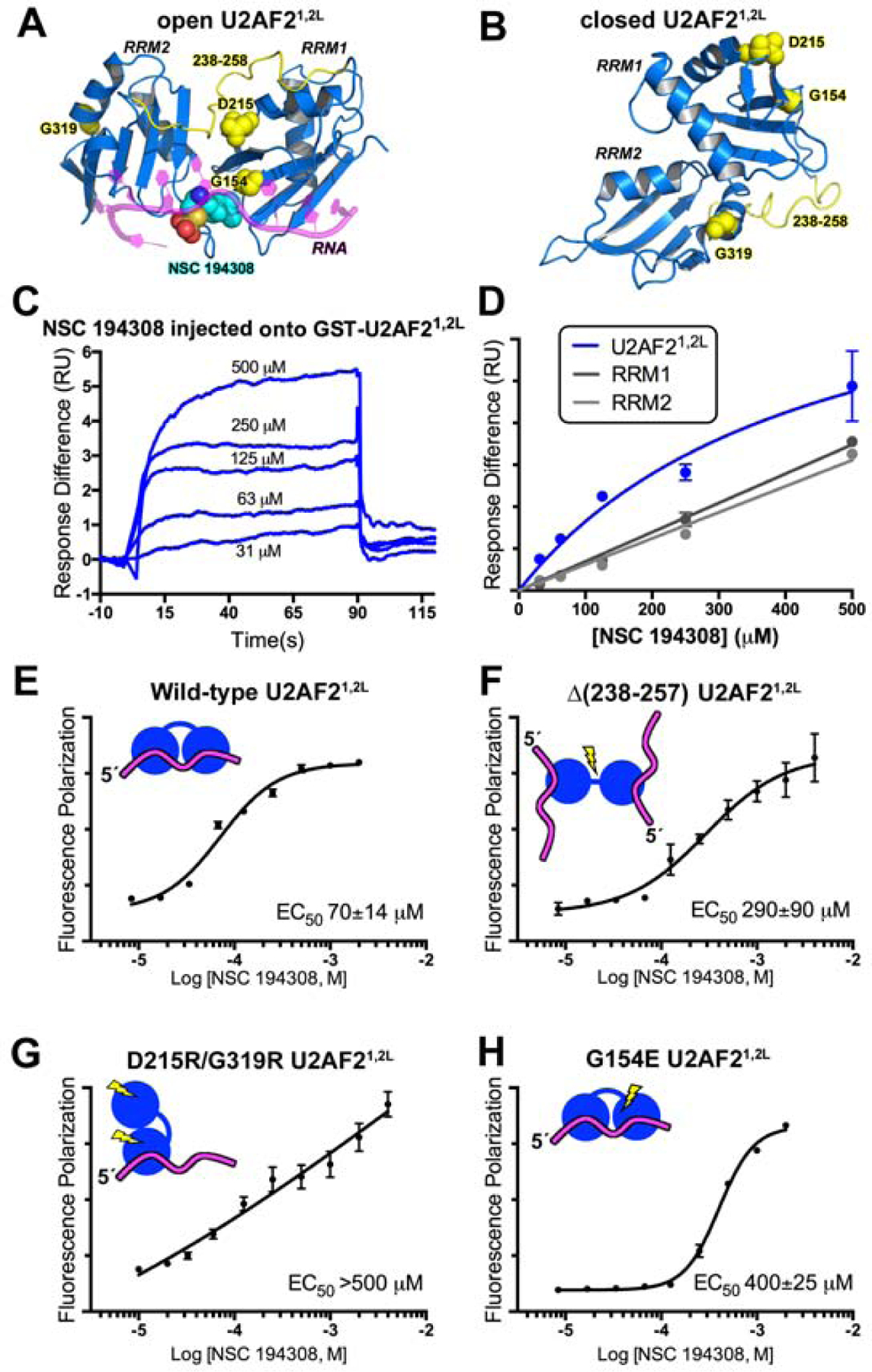
The hit enhancer NSC 194308 binds U2AF2 RRM1/RRM2 (U2AF21,2L). (A) The favorable predicted binding site for NSC 194308 is located between RRM1 and RRM2 (blue) of the open U2AF21,2L conformation (PDB ID 5EV4). Candock scores are listed in Table S2. The bound oligonucleotide (magenta) of the structure is overlaid for reference. Structure-guided mutants are yellow, and for single-site mutations, shown as space-filling CPKs; NSC 194308 is a CPK representation colored by atom: carbon, cyan; oxygen, red; sulfur, orange; nitrogen, blue. (B) Locations of the mutated-regions (yellow) shown on the NMR model (blue) of closed U2AF21,2L (PDB ID 2YH0). (C) Representative sensorgram showing the aligned responses of NSC 194308 injected at the indicated concentrations over an immobilized GST-U2AF21,2L surface. (D) Plot of the average responses (two replicates) from the saturated regions of the sensorgrams fit to a nonlinear steady-state model. NSC 194308 binding to GST-U2AF21,2L, blue; separate GST-RRM1, dark gray; GST-RRM2, light gray. (E-H) Fluorescence polarization dose-responses of the hit enhancer titrated into DEK FLRNA bound to U2AF21,2L variants, including (E) wild-type, (F) inter-RRM linker deletion, (G) D215R/G319R, or (H) G154E. The mean ± SD of three replicates are shown in E - H. Schematic diagrams of the expected conformations of the U2AF2 variants are inset and colored as in A.
