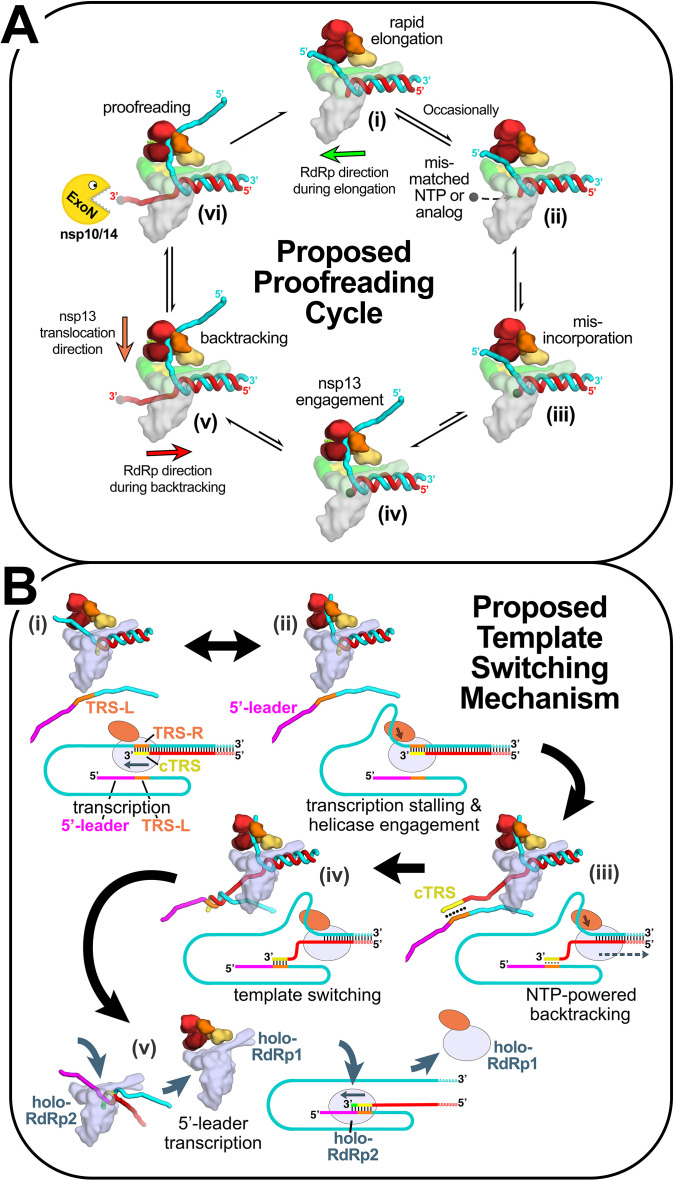
Fig. 7.
Structural basis for the proposed role of nsp13-mediated backtracking during proofreading and template switching during sg-transcription [7], [101]. Structural models are shown as cartoons (holo-RdRp, light blue or gray; nsp13.1 helicase, orange shades; RNA strands, colored tubes). The nsp13.2 helicase is not shown for clarity (all of the models are compatible with the presence of nsp13.2). With each structural diagram is a schematic cartoon illustrating the arrangement of RNA strands. Additional proteins involved in these processes are omitted. The product RNA (p-RNA) being elongated by the RdRp is shown in red. (A) Structural basis for the proposed role of nsp13-mediated backtracking during proofreading: (i) In most cases, the RdRp elongates the p-RNA rapidly and processively [99] (in this orientation, the RdRp moves from right to left; green arrow). The single-stranded 5′-t-RNA segment (cyan) is not engaged with nsp13; (ii), (iii) Infrequently, a mis-matched NTP or a nucleotide analog (represented by the black dot) binds in the RdRp active site (ii) and is incorporated at the p-RNA 3′-end (iii); (iv) The misincorporation causes the RdRp to pause or stall, allowing nsp13 to engage with the single-stranded 5′-t-RNA segment; (v) nsp13 translocation in the 5′ → 3′ direction (orange arrow) and the hindered RdRp elongation activity (due to the mismatch) results in backtracking (backwards motion of the RdRp, red arrow) with extrusion of the p-RNA 3′-end out the RdRp NTP-entry tunnel [101]; (vi) The p-RNA 3′-end is exposed to the nsp10/14 ExoN proofreading activity, resulting in removal of the mis-incorporation and return to the elongating state. (B) Proposed role of nsp13-mediated backtracking in template-switching associated with sg-transcription [see Refs. [4], [14], [83], [102]: (i) (−)-strand RNA synthesis proceeds from the genomic 3′-poly(A)-tail until a Transcription-Regulating Sequence [TRS-R, orange; [15]] is transcribed (cTRS, yellow); (ii) The TRS causes transcription complex stalling; (iii) Helicase function acting on the (+)-strand RNA (cyan) causes backtracking of the transcription complex [101], freeing the pRNA 3′-end; (iv) The p-RNA 3′-end cTRS (yellow) hybridizes with the complementary TRS-L (orange) following the genomic 5′-leader sequence [magenta; [15], [16], [18]; (v) Processive helicase function backtracks the RdRp complex and unwinds the p-RNA from the genomic 3′-end. A second holo-RdRp (holo-RdRp2) can load into the p-RNA 3′-end and continue transcription using the 5′-leader as template.
This figure is adapted from Fig. 6c of Chen, J., B. Malone, E. Llewellyn, M. Grasso, P.M.M. Shelton, P.D.B. Olinares, K. Maruthi, E.T. Eng, H. Vatandaslar, B.T. Chait, T.M. Kapoor, S.A. Darst, and E.A. Campbell, Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell, 2020 182, 6, 1560–1573.