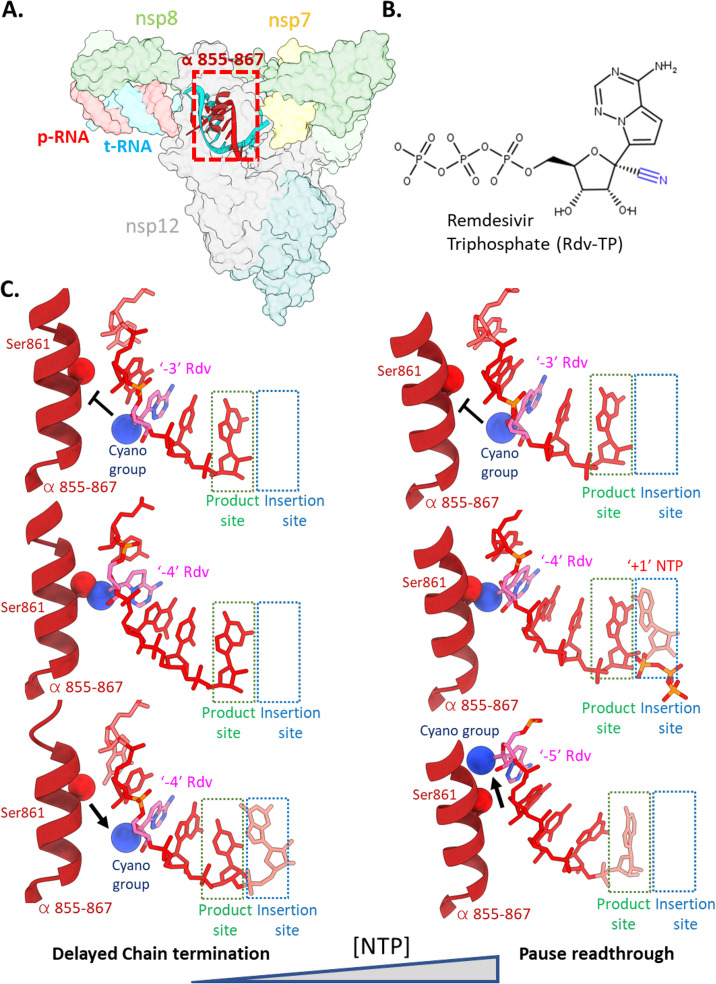
Fig. 9.
(A) The holo-RdRp is the target of Remdesivir (Rdv). (B) Chemical structure of Rdv-triphosphate (Rdv-TP) with the 1-cyano group highlighted in blue. (C) (Left) At low nucleotide concentrations, Rdv inhibits RNA synthesis via the delayed chain termination mechanism [124], [125], [126]. Following incorporation of Rdv at the 3′-end of the RNA p-strand and translocation, two more cycles of NTP addition and translocation can occur rapidly (top panel). A third NTP following the Rdv can be incorporated by the RdRp, but further translocation is hindered by steric clash of the Rdv 1-cyano group with the side chain of nsp12 Ser861 (PDB 7B3B[127]), forcing the RdRp into a pretranslocated stalled state (7B3C [127]). (Right) At high nucleotide concentrations, the presence of a nucleotide substrate in the “insertion” site prohibits the reversion from post to pre-translocated states. The p-RNA can thus be extended past the pause induced by Rdv at the p-RNA “− 3” position [99], [126].