Figure 3.
A quantitative framework to predict gene vulnerability to transcriptional silencing
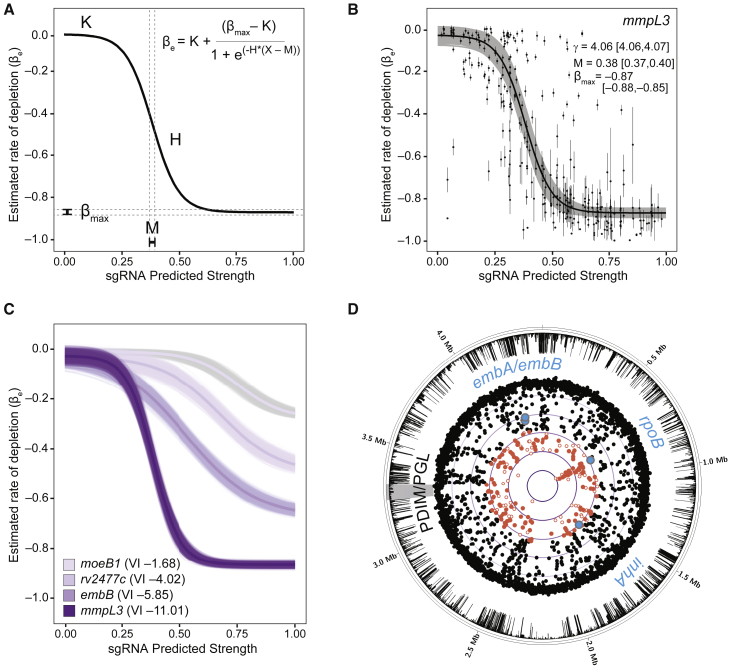
(A) Description of the logistic curve parameters used to model gene level vulnerability. The x axis depicts the linear model predicted strengths of gene-targeting sgRNAs. The y axis depicts the fitness cost of individual sgRNAs estimated by the two-line model from Figure 2A. See details in STAR Methods.
(B) Logistic curve fit to all sgRNAs (dots) targeting mmpL3. The black line represents the mean logistic curve and range (gray) from 5,000 parameter samples. Mean parameter estimates and their 95% highest density interval (HDI) are indicated.
(C) Logistic regression fits for four example genes of differing vulnerability along with their corresponding VI. Lines represent fits generated by the sampling procedure with the dark line representing the mean fit.
(D) Circos plot showing all targeted Mtb H37Rv genes (dots) with their VI. Genes in the upper quartile of vulnerability are depicted as red dots (filled red, confident VI; unfilled red, low-confidence VI). Genes encoding the targets of first line TB therapy (rpoB, inhA, and embAB) are highlighted by blue dots. The outer ring represents the gene-level L2FC value at 28.8 generations. The inner purple lines represent decreasing VI values, with the most vulnerable genes located closest to the center of the circle. The phthiocerol dimycocerosates (PDIM)/phenolic glycolipid (PGL) locus (gray) contains no vulnerable genes.