Figure 2.
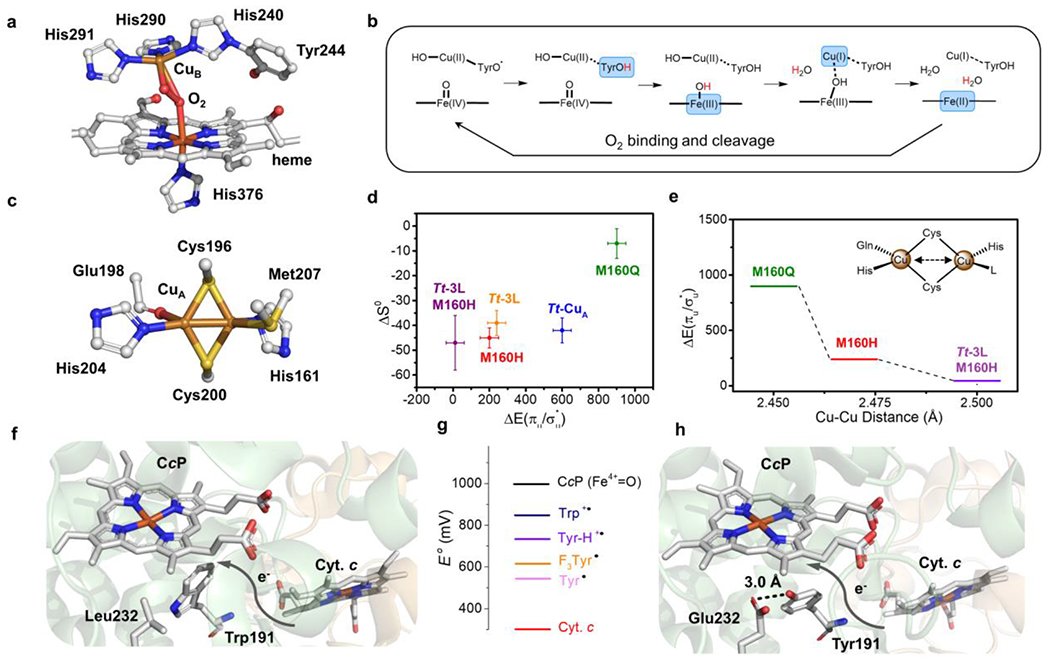
(a) Heme-CuB site (BNC) of bovine heart HCO (PDB 2OCC) with O2 bound. (b) Proposed mechanism for HCO which shows the four-electron reduction from fully oxidized BNC to the rest state. Each arrow on the top represents an one-proton-one-electron reduction. The reduced moieties in each step are showed in blue blocks and the coupled protons are labelled in red. (c) Active site structure of Tt-CuA (PDB 2CUA). (d) Scatter plot of the Cu(II)-Cu(I) entropy change (ΔS0, y-axis) of Tt-CuA mutants versus the energy gap between two thermally accessible ground states (ΔEσu*/πu), x-axis). (e) Change of (ΔEσu*/πu) (y-axis) with different Cu-Cu distances in Tt-CuA mutants. (f) Structural representation of wild-type yeast CcP (co-crystallized with Cyt. c, PDB 2PCC). (g) Scheme of reduction potentials of different amino acid residue radicals and iron species. (h) Structural representation of W191Y/L232E CcP (PDB 6P41). Die hydrogen bond between Glu232 and Tyr191 is shown as a black dash.
