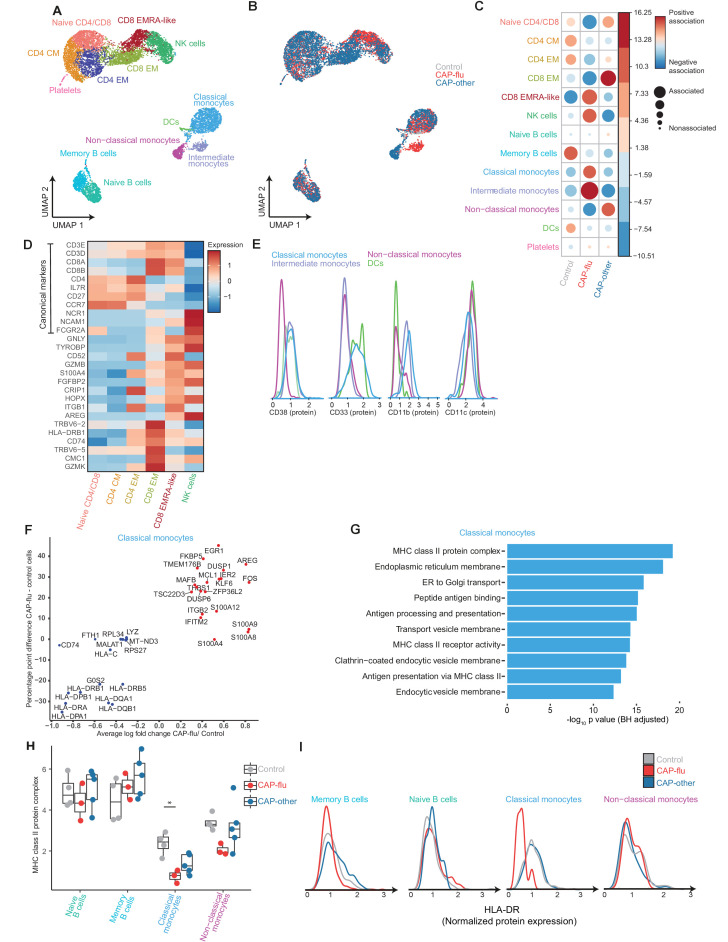
Figure 2. The peripheral immune features of T cells, NK cells, and monocytes vary between CAP-flu and CAP-other.
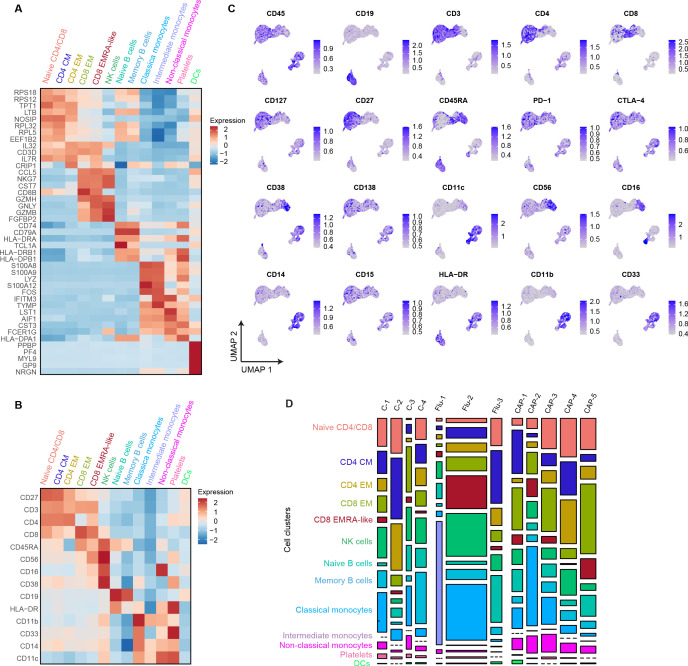
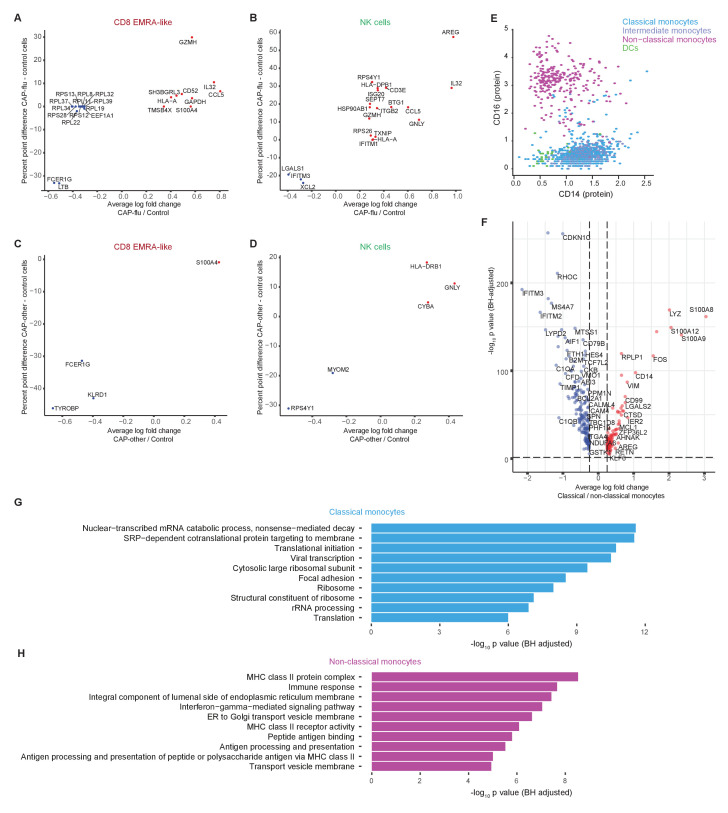
(a, b) UMAPs depicting the clusters identified by the single-cell transcriptomic analysis of PBMCs from controls, CAP-flu, and CAP-other patients, where each dot represents a single cell. In the first UMAP (a), cells are colored by cell type cluster, whereas in the second UMAP (b), cells are colored by donor group. See also Figure 2—figure supplement 1. (c) Correlation plot depicting cluster enrichment in controls, CAP-flu, and CAP-other patients. Dot size proportional to Pearson’s residual of the chi-squared test (i.e., reflecting the difference between the observed and expected proportion), while the color represents the degree of association from Pearson’s chi-squared residuals (red means a positive association, blue means a negative association). (c) Heatmap showing the expression of canonical genes and the top differentially expressed genes (DEGs) derived from comparing the CD8 EM and CD8 EMRA-like cell clusters (adjusted p<0.05). The heatmap also shows the expression of these genes in the other identified T and NK cell clusters. See also Figure 2—figure supplement 2. (e) Density plots showing the surface protein expression of CD38, CD33, CD11b, and CD11c per myeloid cell cluster. (f) Graph depicting the DEGs in the classical monocyte cluster when comparing CAP-flu patients versus controls. The X-axis depicts the average log fold change and the Y-axis depicts the percentage point difference between the proportion of cells expressing the gene in the CAP-flu group minus the proportion of cells expressing the gene in the control group. All depicted DEGs are statistically significant after adjusting for multiple testing (Benjamini-Hochberg [BH]). (g) Bar plot showing Gene Ontology pathway analysis of downregulated genes identified in the analysis in panel (g). The X-axis shows the BH adjusted −log10 p-value from the enrichment score analysis. (h) Boxplots depicting the downregulation of the MHC class II protein complex transcriptional pathway in naive B cells, memory B cells, classical monocytes, and non-classical monocytes clusters, split between controls, CAP-flu, and CAP-other patients. Statistical significance was determined using the two-sided Kruskal-Wallis test with post hoc pairwise Dunn’s test: *BH-adjusted p<0.05. (i) Density plot showing the normalized surface protein expression of HLA-DR on cells in naive B cells, memory B cells, classical monocytes, and non-classical monocytes clusters, split between controls, CAP-flu, and CAP-other patients. CAP, community-acquired pneumonia; NK, natural killer; PBMC, peripheral blood mononuclear cell; UMAP, Uniform Manifold Approximation and Projection.