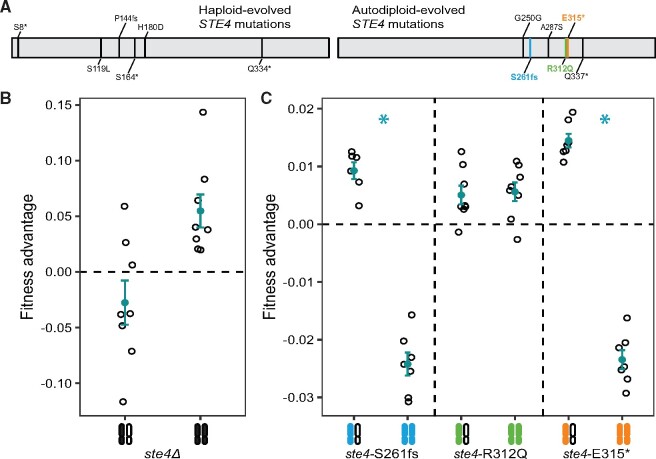
Fig. 1.
Adaptive STE4 mutations differ between haploid and autodiploid populations. (A) Mutations previously identified in the STE4 gene in haploid (Lang et al. 2013) and autodiploid (Fisher et al. 2018) populations. The positions of mutations across the coding sequence of STE4 in haploid populations did not deviate from random expectation (X2(1, N = 6)=0.74363, P = 0.39). Conversely, autodiploid mutations accumulated nonrandomly across the linear 1,276 bp sequence of the STE4 gene, (X2(1, N = 6)=18.76, P < 10−4). Bolded mutations are ones that were selected for reconstruction. (B) Deletion of STE4 is deleterious in autodiploids when heterozygous and beneficial when homozygous. (C) Three evolved autodiploid alleles were reconstructed in an ancestral background. ste4-S261fs and ste4-E315* are overdominant in the ancestral background. (B and C) A filled and open chromosome represents heterozygosity and two filled chromosomes represent homozygosity. Open points are fitness measures of eight biological replicates following MATa/a conversion. Filled points show mean fitness ± SE.