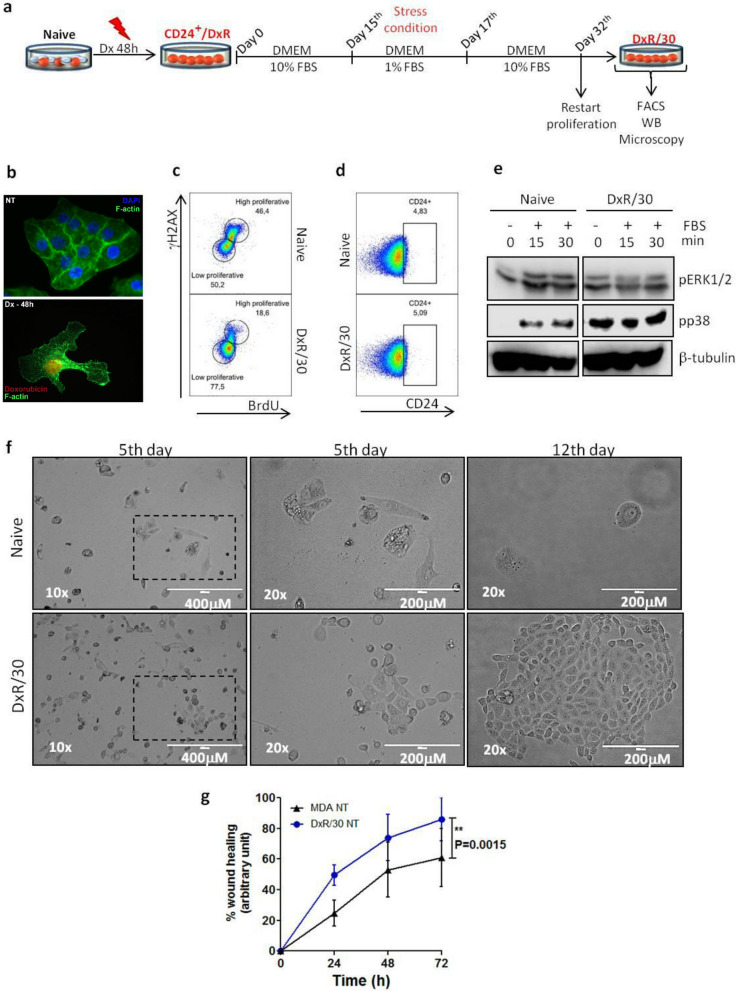
Figure 5.
CD24+/DxR cells become proliferative and more invasive than naïve cells after a long-lasting period in drug-free environment. (a) Schematic shows the journey of CD24+/DxR cells from their switching to CD24+ phenotype to their entry in slow- cycling state and reversion into proliferative cells. CD24+/DxR cells were exposed to a stress condition (fetal bovine serum starvation = 1% FBS) during 48 h. (b) Differences in morphology between naïve MDA-MB-231 and CD24+/DxR cells. Naïve MDA-MB-231 and CD24+/DxR cells were fixed, permeabilized and stained using anti-F-actin/Alexa488 and the nuclear dye DAPI. (c) CD24+/DxR cells retrieve their capacity to proliferate (named as DxR/30 or revertant). Naive MDA-MB-231 and DxR/30 cells were incubated with 10 μM of BrdU (thymine analogue base) for 4 h. Then, cells were stained with anti-BrdU/PerCP-Cy5.5 and anti-γH2AX/Alexa647 (intracellular staining). The low and high proliferative subpopulations were defined and analyzed. The pseudocolor plots are representative of triplicates. (d) DxR/30 cell population returns to the basal ratio of CD24+ cells. Cells were stained with anti-CD24/Pe-Cy7 (extracellular staining). Pseudocolor plots are representative of triplicates. (e) Constitutive activation of p38 MAPK in DxR/30 cells. After starvation for 2 h, cells were stimulated with serum in different times. Cell lysates were immunoblotted with the depicted antibodies. All blots were performed at least twice. (f) Higher resistance of DxR/30 cells to Dx treatment. MDA-MB-231 and DxR/30 cells were treated with Dx (0.6 μM) for 24 h. Then cells were counted, re-seeded in the same density and cultured in normal conditions (DMEM 10% FBS). Cell density and morphology were analyzed at 5 and 12 days post-Dx treatment. (g) DxR/30 cells are more invasive than MDA-MB-231 cells. MDA-MB-231 and DxR/30 cells were seeded (8 × 105) in 6 wells plate and a scratch performed according to Mat and Med. Closing-time percentage was calculated based on the initial scratch size of each cell type. Curves were fitted as means of duplicates ± SD. **p < 0.01 (two-way ANOVA with Bonferroni post-test).