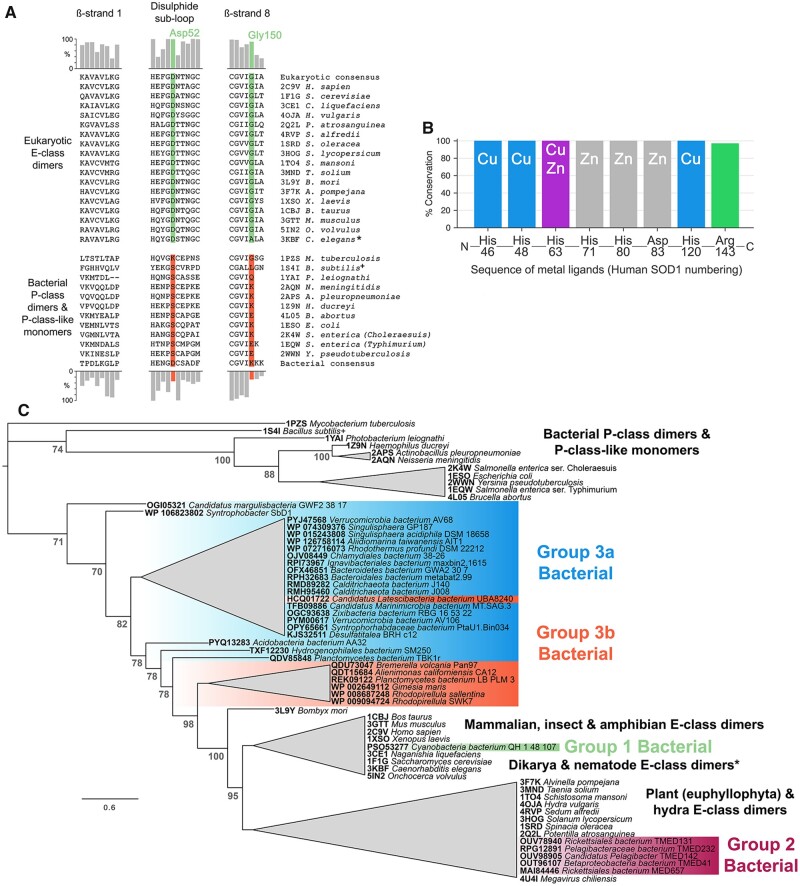
Fig. 1.
Eukaryotic-like CuZnSODs are present in bacteria and can be separated into three groups. (A) Comparison of homodimer interface amino acids found in those bacterial and eukaryotic CuZnSODs represented by structures present in the PDB. Bar charts indicate amino acid conservation with respect to eukaryotic (upper) and bacterial (lower) consensus sequences constructed from 378 eukaryotic and 248 bacterial sequences retrieved from the OMA database (Altenhoff et al. 2018). (B) Bacterial E-class-like CuZnSODs have conserved metal-binding ligand type and order in comparison with other eukaryotic and bacterial enzymes. Catalytically important Arg143 (human numbering) is also very highly conserved with one exception: Candidatus Margulisbacteria bacterium GWF2_38_17 CuZnSOD OGI05321 has proline substitution at this site. (C) Unrooted phylogenetic tree showing eukaryotic-like bacterial CuZnSODs clustered in three groups within the E-class clade rather than with bacterial P-class dimer and P-class-like monomers. Node numbers in gray represent Bayesian posterior probabilities as percentages. Generated using the Jones–Taylor–Thornton substitution model (supplementary fig. S1A, Supplementary Material online). Branches are collapsed where clade topology is unstable across phylogenetic trees generated using different amino acid substitution models (supplementary fig. S1, Supplementary Material online) or where support values are less the 70%. Branch lengths represent expected substitutions per site with a scale bar at the bottom left. * C. elegans CuZnSOD (3KBF) has Ala153 in place of human SOD1 Gly150 and as a result the enzyme is monomeric. + Structure 1S4I from B. subtilis is not an active superoxide dismutase as it cannot bind copper but retains a CuZnSOD-like structure.