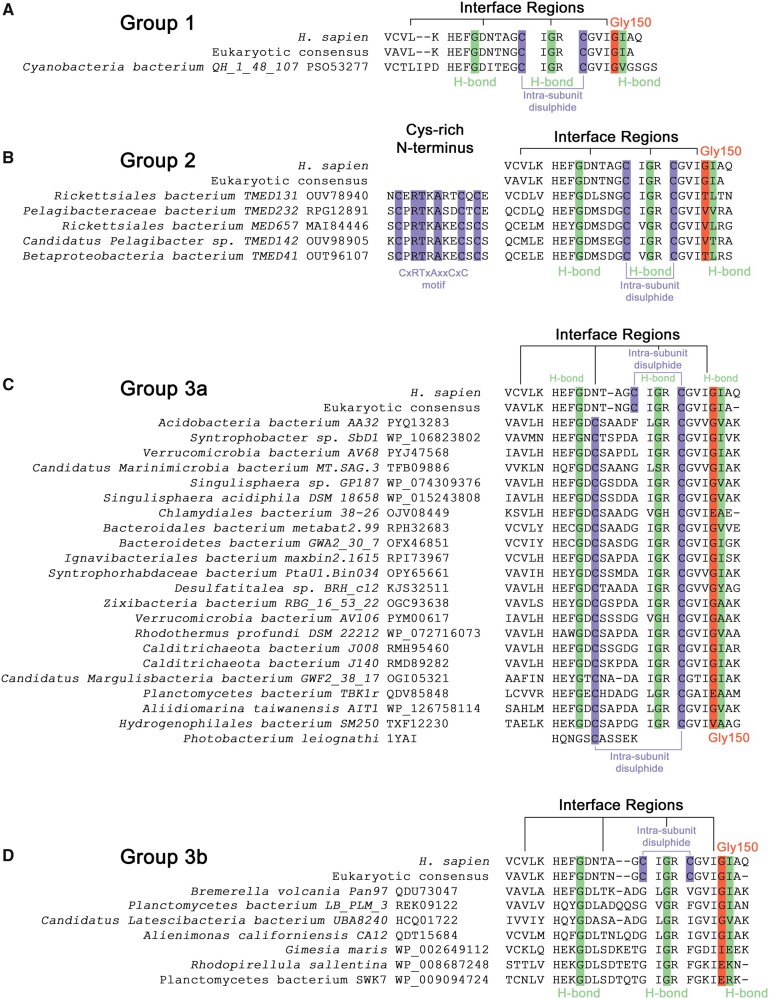
Fig. 2.
Three groups of eukaryotic-like bacterial CuZnSODs have distinct primary structure features. (A) Group 1 Cyanobacteria bacterium QH_1_48_107 CbCuZnSOD has E-class intrasubunit disulfide configuration and interface regions with conservative nonpolar substitutions in interface and intersubunit hydrogen bonding residues. (B) Group 2 enzymes have an N-terminal CxRTxAxxCxC motif, E-class cysteine configuration, conserved hydrogen bonding residues but large and polar substitutions in place of human Gly150 and β-strand 1, respectively. (C) Group 3a interface residues including Gly150 (81% conserved) and intrasubunit hydrogen bonding residues are well conserved from eukaryotic CuZnSODs. Disulfide subloop cysteines involved in intrasubunit disulfide formation are conserved with respect to P-class P. leiognathi CuZnSOD (Bourne et al. 1996). (D) Group 3b enzymes do not have intrasubunit disulfide bonding cysteines. G. maris, R. sallentina, and P. bacterium SWK7 have poorly conserved E-class interface residues in contrast to other members. All include extended N-termini even after removal of signal peptide but do not contain a CXC motif unlike Group 2 enzymes. Green—interface hydrogen bonding residues, orange—Gly150 equivalent residues (human numbering) (Sala et al. 2019), purple—cysteine.