Fig. 7.
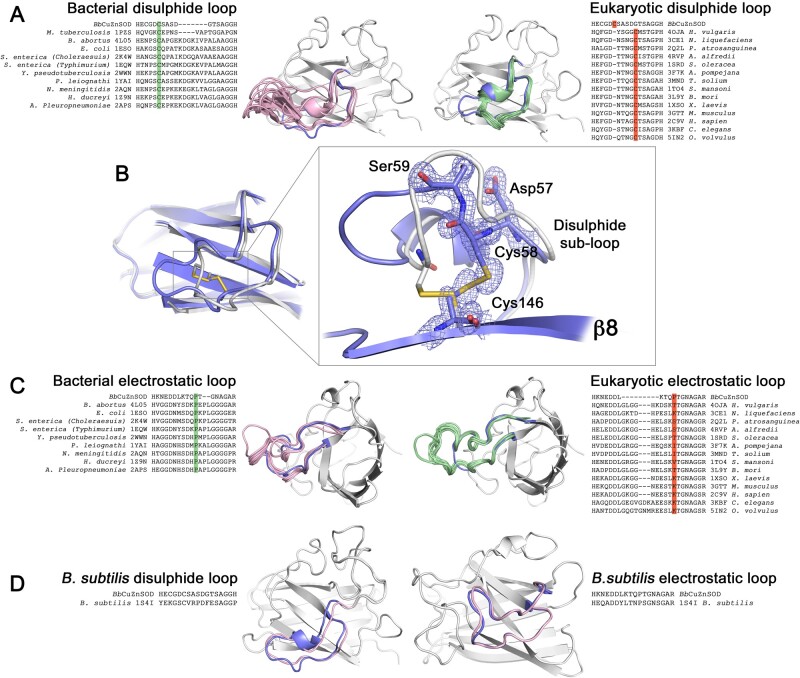
Eukaryotic-like BbCuZnSOD β-barrel loops display similarities to P-class CuZnSODs. (A) Sequence and structural comparison of the BbCuZnSOD disulfide subloop with analogous P-class and E-class CuZnSOD structures shows BbCuZnSOD loops are of intermediate amino acid length and conformational extension. Structural features are highlighted pink—bacterial, green—eukaryotic, and blue—BbCuZnSOD with all structures aligned to the BbCuZnSOD β-barrel (gray). Sequence data show BbCuZnSOD has a bacterial disulfide subloop cysteine configuration (green) as opposed to eukaryotic (orange). (B) 2Fo-Fc electron density map contoured at 2σ showing BbCuZnSOD has a characteristically prokaryotic intrasubunit disulfide bond configuration (blue) in comparison with human SOD1 (gray). (C) Sequence and structural comparison of the BbCuZnSOD electrostatic loop with bacterial and eukaryotic structures. The BbCuZnSOD has length and conformation similar to bacterial orthologs with a characteristic and highly conserved central proline (green) that is absent in eukaryotic enzymes (orange). Structural features are colored as in (A). (D) Comparison of BbCuZnSOD loop sequence and conformation with that of B. subtilis (1S4I). BbCuZnSOD loop identity to B. subtilis is 47% and 56% in comparison with Alvinella pompejana 59% and 61% for disulfide subloop and electrostatic loops, respectively (gaps excluded). Structural features are colored as in (A).