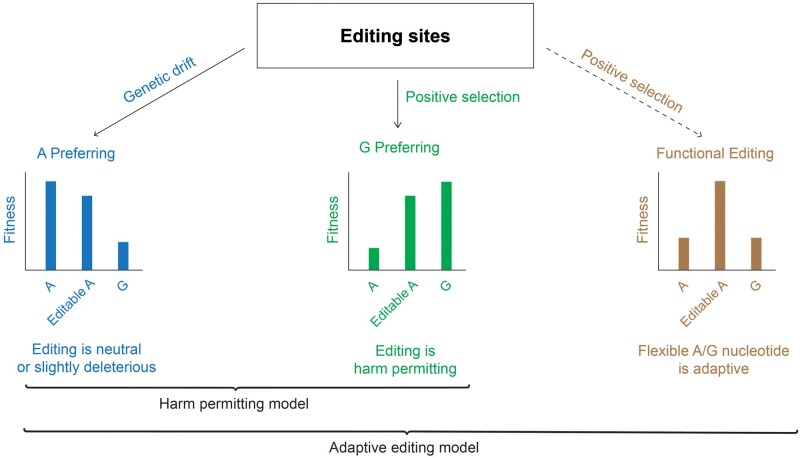
Fig. 1.
The harm-permitting and adaptive editing models. Recoding sites may be fixated into the genome due to random genomic drift, even though their editing does not provide any selective advantage and may be even slightly deleterious. These randomly fixated sites (A-preferring sites) are not expected to be enriched in nonsynonymous editing (left). In a second class of recoding sites, editing does provide a selective advantage as it replaces an inferior A allele by the preferred G allele (middle). For these G-preferring sites, editing does increase the fitness of the organism over an uneditable A, but having a genomically encoded G is equally beneficial or even better (middle). In both cases, fitness does not depend on the protein diversity and flexibility provided by recoding. The HPM asserts that all (or almost all) recoding sites belong to these two categories. In contrast, according to the adaptive editing model some of the recoding sites belong to a third class, where having a recodable codon is functionally important, and the editable A provides a selective advantage over both an uneditable A and a genomically encoded G (right).