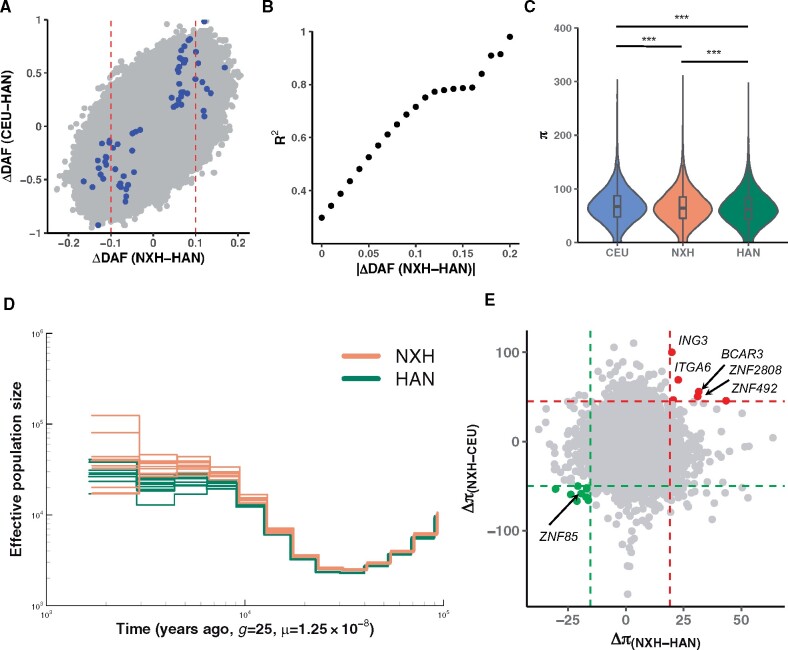
Fig. 5.
The impact of admixture on the genome diversity of NXH. (A) The correlation between allele frequency difference between NXH and HAN and that between western ancestry (CEU) and eastern ancestry (HAN). The red line indicates the top 1% threshold of the empirical distribution of the absolute difference between NXH and HAN. The colored points indicate they are highly differentiated functional variants between NXH and HAN. (B) The x axis indicates the threshold which is used to prune the data set. SNPs with absolute allele frequency larger than the threshold were used to estimate the correlation. The y axis indicates the correlation (measured by R2) between allele frequency difference between NXH and HAN and that between CEU and HAN for the pruned data set. (C) The nucleotide diversity (π) of CEU, NXH, and HAN. An equal number of samples were selected from each population. The “***” indicates P < 0.001. (D) Estimated changes in historical effective population size. Four genomes (eight haploid genomes) were randomly selected from one of the two populations with deeply sequenced genomes in this study. This analysis was repeated 10 times. We showed the results based on an absolute estimation of time under the assumption of a slow mutation rate of 0.5 × 10−9 per site per year. (E) The difference in nucleotide diversity (π) between NXH and HAN and between NXH and CEU populations. Each point represents one nonoverlap 100 kb window on the genome. The dashed line indicates the difference of this window is significant with P < 0.001 of the empirical distribution. Gene names from the significant windows are labeled on the plot.