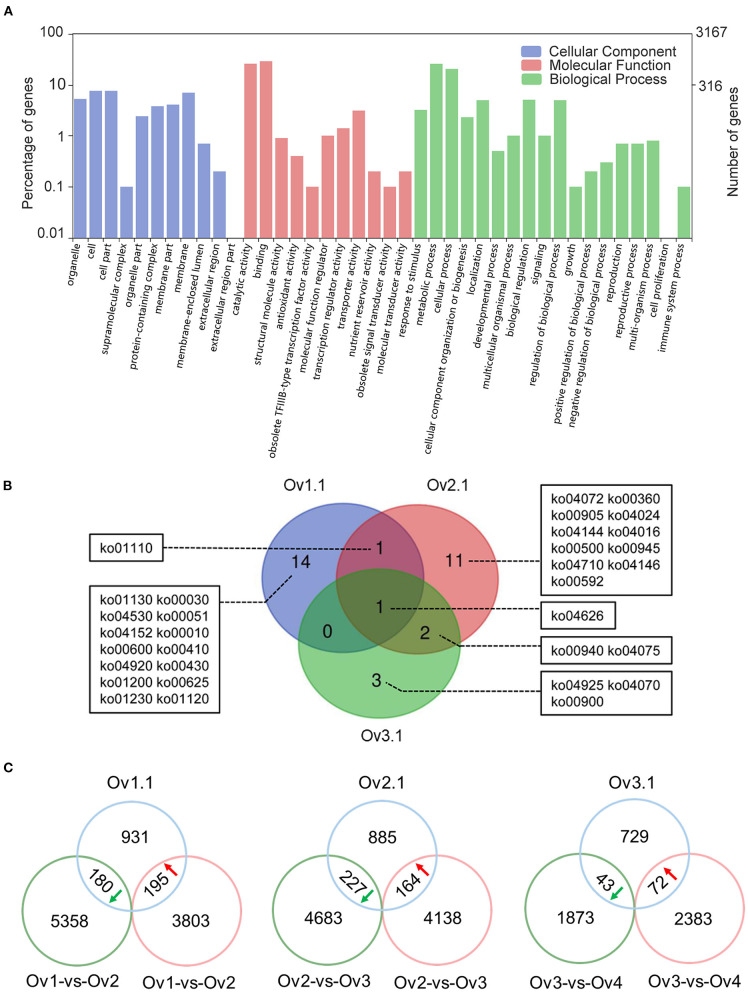
Figure 6.
GO and KEGG enrichment and ChARF3-regulated genes analysis. (A) GO classification of all ChARF3 target genes. (B) Overlaps of significantly enriched KEGG pathways at three ovule development stages. ko00010: Glycolysis/Gluconeogenesis, ko00030: Pentose phosphate pathway, ko00051: Fructose and mannose metabolism, ko00360: Phenylalanine metabolism, ko00410: beta-Alanine metabolism, ko00430: Taurine and hypotaurine metabolism, ko00500: Starch and sucrose metabolism, ko00592: alpha-Linolenic acid metabolism, ko00600: Sphingolipid metabolism, ko00625: Chloroalkane and chloroalkene degradation, ko00900: Terpenoid backbone biosynthesis, ko00905: Brassinosteroid biosynthesis, ko00940: Phenylpropanoid biosynthesis, ko00945: Stilbenoid, diarylheptanoid and gingerol biosynthesis, ko01110: Biosynthesis of secondary metabolites, ko01120: Microbial metabolism in diverse environments, ko01130: Biosynthesis of antibiotics, ko01200: Carbon metabolism, ko01230: Biosynthesis of amino acids, ko04016: MAPK signaling pathway—plant, ko04024: cAMP signaling pathway, ko04070: Phosphatidylinositol signaling system, ko04072: Phospholipase D signaling pathway, ko04075: Plant hormone signal transduction, ko04144: Endocytosis, ko04146: Peroxisome, ko04152: AMPK signaling pathway, ko04530: Tight junction, ko04626: Plant-pathogen interaction, ko04710: Circadian rhythm, ko04920: Adipocytokine signaling pathway, ko04925: Aldosterone synthesis and secretion. (C) Overlap between ChIP-Seq target genes and RNA-Seq DEGs. Green and red circles refer to down- and up-regulated differentially expressed genes in paired comparison respectively, and blue circle refers to genes bound by ChARF3 through ChIP-seq.