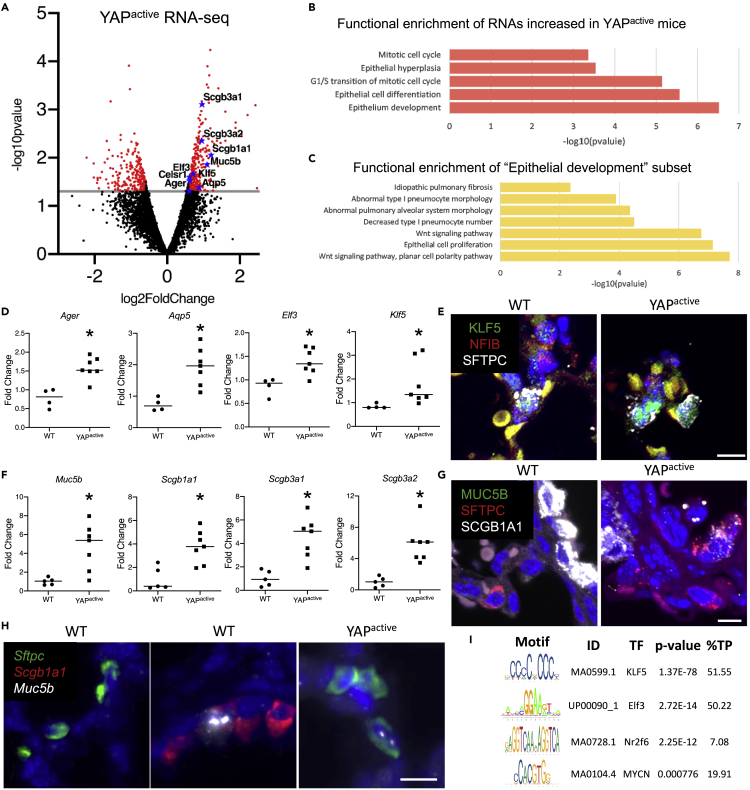
Figure 6.
YAP activation increases expression of AT1 and proximal epithelial cell signature genes
RNA-seq analyses were performed on EPCAM+ cells isolated from D14 YAPactive (N = 6) and controls (N = 4) mice.
(A) Volcano plot of the differentially expressed genes (p < .05, FC > 1.5) shows increase in AT1 markers Ager and Aqp5 as well as proximal lung airway markers Muc5b, Scgb1a1, Scgb3a1 and Scgb3a2.
(B) Genes upregulated in the YAPactive epithelium are associated with mitotic cell cycle and epithelial cell differentiation and development.
(C) Functional enrichment analyses of the subset of genes associated with epithelium development revealed increases in the expression of genes associated with Wnt signaling, epithelium proliferation, IPF and abnormal AT1 differentiation.
(D) Realtime qRT-PCR analyses of isolated YAPactive epithelial cells demonstrate increased Ager, Aqp5, Elf3 and Klf5. ∗Indicates p<0.05 as determined by Welch's t-test.
(E) Immunofluorescence analysis of NFIB (red), KLF5 (green), and SFTPC (white) shows presence of KLF5 in SFTPC+ cells including the SFTPC+ AT2 cell doublets identified in YAPactive mice.
(F) qRT-PCR analysis of proximal cell signature genes demonstrate increased conducting airway epithelial cell markers in YAPactive epithelial cells. ∗Indicates p<0.05 as determined by Welch's t-test.
(G) Immunofluorescence analysis of MUC5B (green), SFTPC (red), and SCGB1A1 (white), shows the presence of SCGB1A1+/SFTPC+ cells in YAPactive mice.
(H) RNAscope Fluorescent in-situ hybridization of Muc5b, Scgb1a1, and Sftpc in WT and YAPactive mouse lungs demonstrating the presence of Sftpc+/Muc5b+ cells in YAPactive lungs.
(I) Promoter analyses of the genes increased in the YAPactive epithelium show that transcription factors Klf5, Elf3, Nr2f6, and Mycn RNAs are increased and have enriched binding sites in YAPactive induced genes. See Table S1 for pathways down regulated in YAPactive mice and Figure S4 for immunofluorescence images.